Naimul Khan
Ryerson University, Toronto, Canada
DynaGuide: A Generalizable Dynamic Guidance Framework for Unsupervised Semantic Segmentation
Feb 13, 2026Abstract:Unsupervised image segmentation is a critical task in computer vision. It enables dense scene understanding without human annotations, which is especially valuable in domains where labelled data is scarce. However, existing methods often struggle to reconcile global semantic structure with fine-grained boundary accuracy. This paper introduces DynaGuide, an adaptive segmentation framework that addresses these challenges through a novel dual-guidance strategy and dynamic loss optimization. Building on our previous work, DynaSeg, DynaGuide combines global pseudo-labels from zero-shot models such as DiffSeg or SegFormer with local boundary refinement using a lightweight CNN trained from scratch. This synergy allows the model to correct coarse or noisy global predictions and produce high-precision segmentations. At the heart of DynaGuide is a multi-component loss that dynamically balances feature similarity, Huber-smoothed spatial continuity, including diagonal relationships, and semantic alignment with the global pseudo-labels. Unlike prior approaches, DynaGuide trains entirely without ground-truth labels in the target domain and supports plug-and-play integration of diverse guidance sources. Extensive experiments on BSD500, PASCAL VOC2012, and COCO demonstrate that DynaGuide achieves state-of-the-art performance, improving mIoU by 17.5% on BSD500, 3.1% on PASCAL VOC2012, and 11.66% on COCO. With its modular design, strong generalization, and minimal computational footprint, DynaGuide offers a scalable and practical solution for unsupervised segmentation in real-world settings. Code available at: https://github.com/RyersonMultimediaLab/DynaGuide
Beyond Identity: A Generalizable Approach for Deepfake Audio Detection
May 10, 2025Abstract:Deepfake audio presents a growing threat to digital security, due to its potential for social engineering, fraud, and identity misuse. However, existing detection models suffer from poor generalization across datasets, due to implicit identity leakage, where models inadvertently learn speaker-specific features instead of manipulation artifacts. To the best of our knowledge, this is the first study to explicitly analyze and address identity leakage in the audio deepfake detection domain. This work proposes an identity-independent audio deepfake detection framework that mitigates identity leakage by encouraging the model to focus on forgery-specific artifacts instead of overfitting to speaker traits. Our approach leverages Artifact Detection Modules (ADMs) to isolate synthetic artifacts in both time and frequency domains, enhancing cross-dataset generalization. We introduce novel dynamic artifact generation techniques, including frequency domain swaps, time domain manipulations, and background noise augmentation, to enforce learning of dataset-invariant features. Extensive experiments conducted on ASVspoof2019, ADD 2022, FoR, and In-The-Wild datasets demonstrate that the proposed ADM-enhanced models achieve F1 scores of 0.230 (ADD 2022), 0.604 (FoR), and 0.813 (In-The-Wild), consistently outperforming the baseline. Dynamic Frequency Swap proves to be the most effective strategy across diverse conditions. These findings emphasize the value of artifact-based learning in mitigating implicit identity leakage for more generalizable audio deepfake detection.
Temporal Feature Weaving for Neonatal Echocardiographic Viewpoint Video Classification
Jan 07, 2025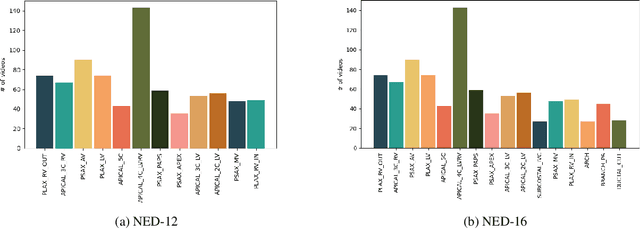


Abstract:Automated viewpoint classification in echocardiograms can help under-resourced clinics and hospitals in providing faster diagnosis and screening when expert technicians may not be available. We propose a novel approach towards echocardiographic viewpoint classification. We show that treating viewpoint classification as video classification rather than image classification yields advantage. We propose a CNN-GRU architecture with a novel temporal feature weaving method, which leverages both spatial and temporal information to yield a 4.33\% increase in accuracy over baseline image classification while using only four consecutive frames. The proposed approach incurs minimal computational overhead. Additionally, we publish the Neonatal Echocardiogram Dataset (NED), a professionally-annotated dataset providing sixteen viewpoints and associated echocardipgraphy videos to encourage future work and development in this field. Code available at: https://github.com/satchelfrench/NED
Enhanced Cross-Dataset Electroencephalogram-based Emotion Recognition using Unsupervised Domain Adaptation
Nov 19, 2024Abstract:Emotion recognition has significant potential in healthcare and affect-sensitive systems such as brain-computer interfaces (BCIs). However, challenges such as the high cost of labeled data and variability in electroencephalogram (EEG) signals across individuals limit the applicability of EEG-based emotion recognition models across domains. These challenges are exacerbated in cross-dataset scenarios due to differences in subject demographics, recording devices, and presented stimuli. To address these issues, we propose a novel approach to improve cross-domain EEG-based emotion classification. Our method, Gradual Proximity-guided Target Data Selection (GPTDS), incrementally selects reliable target domain samples for training. By evaluating their proximity to source clusters and the models confidence in predicting them, GPTDS minimizes negative transfer caused by noisy and diverse samples. Additionally, we introduce Prediction Confidence-aware Test-Time Augmentation (PC-TTA), a cost-effective augmentation technique. Unlike traditional TTA methods, which are computationally intensive, PC-TTA activates only when model confidence is low, improving inference performance while drastically reducing computational costs. Experiments on the DEAP and SEED datasets validate the effectiveness of our approach. When trained on DEAP and tested on SEED, our model achieves 67.44% accuracy, a 7.09% improvement over the baseline. Conversely, training on SEED and testing on DEAP yields 59.68% accuracy, a 6.07% improvement. Furthermore, PC-TTA reduces computational time by a factor of 15 compared to traditional TTA methods. Our method excels in detecting both positive and negative emotions, demonstrating its practical utility in healthcare applications. Code available at: https://github.com/RyersonMultimediaLab/EmotionRecognitionUDA
DynaSeg: A Deep Dynamic Fusion Method for Unsupervised Image Segmentation Incorporating Feature Similarity and Spatial Continuity
May 09, 2024Abstract:Our work tackles the fundamental challenge of image segmentation in computer vision, which is crucial for diverse applications. While supervised methods demonstrate proficiency, their reliance on extensive pixel-level annotations limits scalability. In response to this challenge, we present an enhanced unsupervised Convolutional Neural Network (CNN)-based algorithm called DynaSeg. Unlike traditional approaches that rely on a fixed weight factor to balance feature similarity and spatial continuity, requiring manual adjustments, our novel, dynamic weighting scheme automates parameter tuning, adapting flexibly to image details. We also introduce the novel concept of a Silhouette Score Phase that addresses the challenge of dynamic clustering during iterations. Additionally, our methodology integrates both CNN-based and pre-trained ResNet feature extraction, offering a comprehensive and adaptable approach. We achieve state-of-the-art results on diverse datasets, with a notable 12.2% and 14.12% mIOU improvement compared to the current benchmarks on COCO-All and COCO-Stuff, respectively. The proposed approach unlocks the potential for unsupervised image segmentation and addresses scalability concerns in real-world scenarios by obviating the need for meticulous parameter tuning.
A Dynamically Weighted Loss Function for Unsupervised Image Segmentation
Mar 17, 2024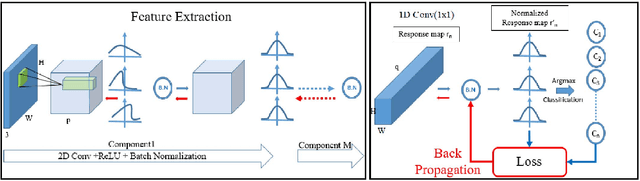

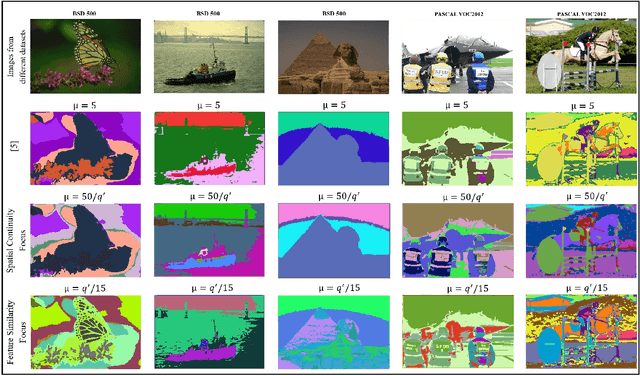
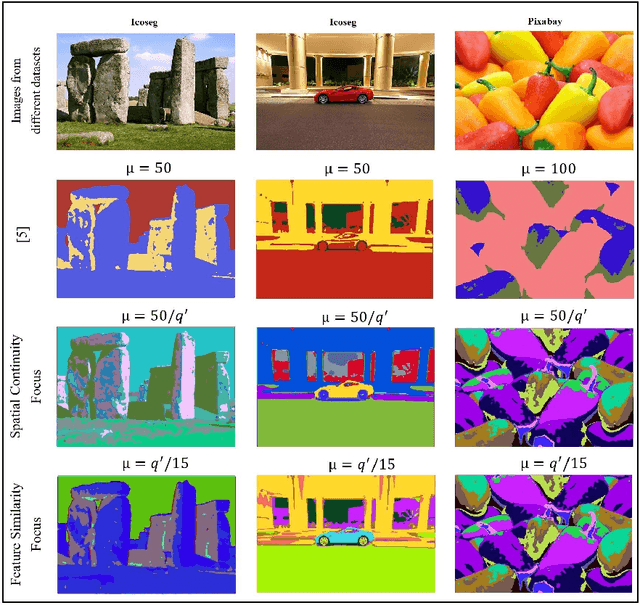
Abstract:Image segmentation is the foundation of several computer vision tasks, where pixel-wise knowledge is a prerequisite for achieving the desired target. Deep learning has shown promising performance in supervised image segmentation. However, supervised segmentation algorithms require a massive amount of data annotated at a pixel level, thus limiting their applicability and scalability. Therefore, there is a need to invest in unsupervised learning for segmentation. This work presents an improved version of an unsupervised Convolutional Neural Network (CNN) based algorithm that uses a constant weight factor to balance between the segmentation criteria of feature similarity and spatial continuity, and it requires continuous manual adjustment of parameters depending on the degree of detail in the image and the dataset. In contrast, we propose a novel dynamic weighting scheme that leads to a flexible update of the parameters and an automatic tuning of the balancing weight between the two criteria above to bring out the details in the images in a genuinely unsupervised manner. We present quantitative and qualitative results on four datasets, which show that the proposed scheme outperforms the current unsupervised segmentation approaches without requiring manual adjustment.
Cross-Database and Cross-Channel ECG Arrhythmia Heartbeat Classification Based on Unsupervised Domain Adaptation
Jun 07, 2023



Abstract:The classification of electrocardiogram (ECG) plays a crucial role in the development of an automatic cardiovascular diagnostic system. However, considerable variances in ECG signals between individuals is a significant challenge. Changes in data distribution limit cross-domain utilization of a model. In this study, we propose a solution to classify ECG in an unlabeled dataset by leveraging knowledge obtained from labeled source domain. We present a domain-adaptive deep network based on cross-domain feature discrepancy optimization. Our method comprises three stages: pre-training, cluster-centroid computing, and adaptation. In pre-training, we employ a Distributionally Robust Optimization (DRO) technique to deal with the vanishing worst-case training loss. To enhance the richness of the features, we concatenate three temporal features with the deep learning features. The cluster computing stage involves computing centroids of distinctly separable clusters for the source using true labels, and for the target using confident predictions. We propose a novel technique to select confident predictions in the target domain. In the adaptation stage, we minimize compacting loss within the same cluster, separating loss across different clusters, inter-domain cluster discrepancy loss, and running combined loss to produce a domain-robust model. Experiments conducted in both cross-domain and cross-channel paradigms show the efficacy of the proposed method. Our method achieves superior performance compared to other state-of-the-art approaches in detecting ventricular ectopic beats (V), supraventricular ectopic beats (S), and fusion beats (F). Our method achieves an average improvement of 11.78% in overall accuracy over the non-domain-adaptive baseline method on the three test datasets.
Structure Preserving Cycle-GAN for Unsupervised Medical Image Domain Adaptation
Apr 18, 2023Abstract:The presence of domain shift in medical imaging is a common issue, which can greatly impact the performance of segmentation models when dealing with unseen image domains. Adversarial-based deep learning models, such as Cycle-GAN, have become a common model for approaching unsupervised domain adaptation of medical images. These models however, have no ability to enforce the preservation of structures of interest when translating medical scans, which can lead to potentially poor results for unsupervised domain adaptation within the context of segmentation. This work introduces the Structure Preserving Cycle-GAN (SP Cycle-GAN), which promotes medical structure preservation during image translation through the enforcement of a segmentation loss term in the overall Cycle-GAN training process. We demonstrate the structure preserving capability of the SP Cycle-GAN both visually and through comparison of Dice score segmentation performance for the unsupervised domain adaptation models. The SP Cycle-GAN is able to outperform baseline approaches and standard Cycle-GAN domain adaptation for binary blood vessel segmentation in the STARE and DRIVE datasets, and multi-class Left Ventricle and Myocardium segmentation in the multi-modal MM-WHS dataset. SP Cycle-GAN achieved a state of the art Myocardium segmentation Dice score (DSC) of 0.7435 for the MR to CT MM-WHS domain adaptation problem, and excelled in nearly all categories for the MM-WHS dataset. SP Cycle-GAN also demonstrated a strong ability to preserve blood vessel structure in the DRIVE to STARE domain adaptation problem, achieving a 4% DSC increase over a default Cycle-GAN implementation.
Lightweight and Interpretable Left Ventricular Ejection Fraction Estimation using Mobile U-Net
Apr 17, 2023



Abstract:Accurate LVEF measurement is important in clinical practice as it identifies patients who may be in need of life-prolonging treatments. This paper presents a deep learning based framework to automatically estimate left ventricular ejection fraction from an entire 4-chamber apical echocardiogram video. The aim of the proposed framework is to provide an interpretable and computationally effective ejection fraction prediction pipeline. A lightweight Mobile U-Net based network is developed to segment the left ventricle in each frame of an echocardiogram video. An unsupervised LVEF estimation algorithm is implemented based on Simpson's mono-plane method. Experimental results on a large public dataset show that our proposed approach achieves comparable accuracy to the state-of-the-art while being significantly more space and time efficient (with 5 times fewer parameters and 10 times fewer FLOPS).
Classification of Lung Pathologies in Neonates using Dual Tree Complex Wavelet Transform
Feb 17, 2023Abstract:Annually 8500 neonatal deaths are reported in the US due to respiratory failure. Recently, Lung Ultrasound (LUS), due to its radiation free nature, portability, and being cheaper is gaining wide acceptability as a diagnostic tool for lung conditions. However, lack of highly trained medical professionals has limited its use especially in remote areas. To address this, an automated screening system that captures characteristics of the LUS patterns can be of significant assistance to clinicians who are not experts in lung ultrasound (LUS) images. In this paper, we propose a feature extraction method designed to quantify the spatially-localized line patterns and texture patterns found in LUS images. Using the dual-tree complex wavelet transform (DTCWT) and four types of common image features we propose a method to classify the LUS images into 6 common neonatal lung conditions. These conditions are normal lung, pneumothorax (PTX), transient tachypnea of the newborn (TTN), respiratory distress syndrome (RDS), chronic lung disease (CLD) and consolidation (CON) that could be pneumonia or atelectasis. The proposed method using DTCWT decomposition extracted global statistical, grey-level co-occurrence matrix (GLCM), grey-level run length matrix (GLRLM) and linear binary pattern (LBP) features to be fed to a linear discriminative analysis (LDA) based classifier. Using 15 best DTCWT features along with 3 clinical features the proposed approach achieved a per-image classification accuracy of 92.78% with a balanced dataset containing 720 images from 24 patients and 74.39% with the larger unbalanced dataset containing 1550 images from 42 patients. Likewise, the proposed method achieved a maximum per-subject classification accuracy of 81.53% with 43 DTCWT features and 3 clinical features using the balanced dataset and 64.97% with 13 DTCWT features and 3 clinical features using the unbalanced dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge