Adel Mohamed
Mount Sinai Hospital, University of Toronto, Toronto, Canada
Classification of Lung Pathologies in Neonates using Dual Tree Complex Wavelet Transform
Feb 17, 2023Abstract:Annually 8500 neonatal deaths are reported in the US due to respiratory failure. Recently, Lung Ultrasound (LUS), due to its radiation free nature, portability, and being cheaper is gaining wide acceptability as a diagnostic tool for lung conditions. However, lack of highly trained medical professionals has limited its use especially in remote areas. To address this, an automated screening system that captures characteristics of the LUS patterns can be of significant assistance to clinicians who are not experts in lung ultrasound (LUS) images. In this paper, we propose a feature extraction method designed to quantify the spatially-localized line patterns and texture patterns found in LUS images. Using the dual-tree complex wavelet transform (DTCWT) and four types of common image features we propose a method to classify the LUS images into 6 common neonatal lung conditions. These conditions are normal lung, pneumothorax (PTX), transient tachypnea of the newborn (TTN), respiratory distress syndrome (RDS), chronic lung disease (CLD) and consolidation (CON) that could be pneumonia or atelectasis. The proposed method using DTCWT decomposition extracted global statistical, grey-level co-occurrence matrix (GLCM), grey-level run length matrix (GLRLM) and linear binary pattern (LBP) features to be fed to a linear discriminative analysis (LDA) based classifier. Using 15 best DTCWT features along with 3 clinical features the proposed approach achieved a per-image classification accuracy of 92.78% with a balanced dataset containing 720 images from 24 patients and 74.39% with the larger unbalanced dataset containing 1550 images from 42 patients. Likewise, the proposed method achieved a maximum per-subject classification accuracy of 81.53% with 43 DTCWT features and 3 clinical features using the balanced dataset and 64.97% with 13 DTCWT features and 3 clinical features using the unbalanced dataset.
An interpretable object detection based model for the diagnosis of neonatal lung diseases using Ultrasound images
May 21, 2021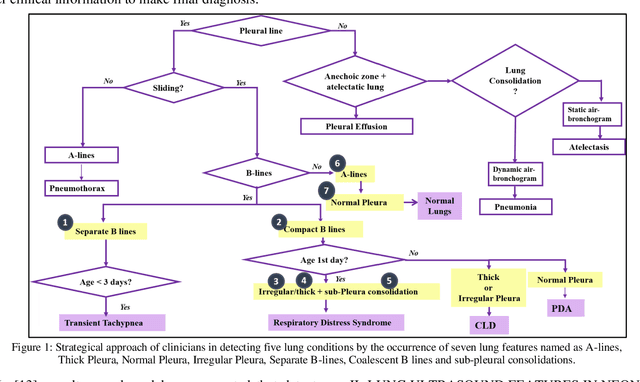
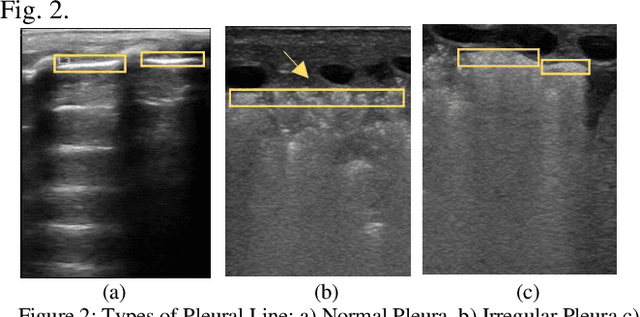
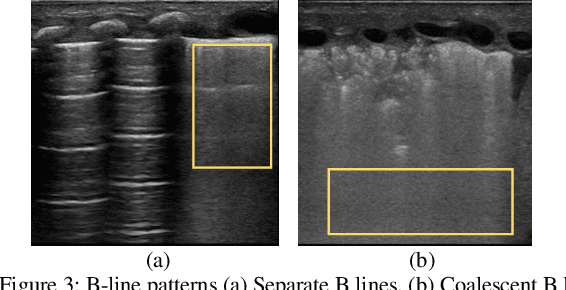
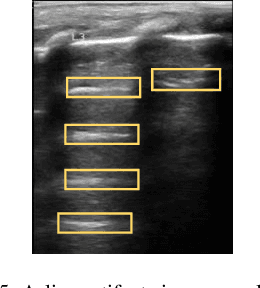
Abstract:Over the last few decades, Lung Ultrasound (LUS) has been increasingly used to diagnose and monitor different lung diseases in neonates. It is a non invasive tool that allows a fast bedside examination while minimally handling the neonate. Acquiring a LUS scan is easy, but understanding the artifacts concerned with each respiratory disease is challenging. Mixed artifact patterns found in different respiratory diseases may limit LUS readability by the operator. While machine learning (ML), especially deep learning can assist in automated analysis, simply feeding the ultrasound images to an ML model for diagnosis is not enough to earn the trust of medical professionals. The algorithm should output LUS features that are familiar to the operator instead. Therefore, in this paper we present a unique approach for extracting seven meaningful LUS features that can be easily associated with a specific pathological lung condition: Normal pleura, irregular pleura, thick pleura, Alines, Coalescent B-lines, Separate B-lines and Consolidations. These artifacts can lead to early prediction of infants developing later respiratory distress symptoms. A single multi-class region proposal-based object detection model faster-RCNN (fRCNN) was trained on lower posterior lung ultrasound videos to detect these LUS features which are further linked to four common neonatal diseases. Our results show that fRCNN surpasses single stage models such as RetinaNet and can successfully detect the aforementioned LUS features with a mean average precision of 86.4%. Instead of a fully automatic diagnosis from images without any interpretability, detection of such LUS features leave the ultimate control of diagnosis to the clinician, which can result in a more trustworthy intelligent system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge