Christopher Nguyen
Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA, USA, Cardiovascular Research Center, Cardiology Division, Massachusetts General Hospital, Charlestown, MA, USA, Health Science Technology, Harvard-MIT, Cambridge, MA, USA
SemiKong: Curating, Training, and Evaluating A Semiconductor Industry-Specific Large Language Model
Nov 22, 2024



Abstract:Large Language Models (LLMs) have demonstrated the potential to address some issues within the semiconductor industry. However, they are often general-purpose models that lack the specialized knowledge needed to tackle the unique challenges of this sector, such as the intricate physics and chemistry of semiconductor devices and processes. SemiKong, the first industry-specific LLM for the semiconductor domain, provides a foundation that can be used to develop tailored proprietary models. With SemiKong 1.0, we aim to develop a foundational model capable of understanding etching problems at an expert level. Our key contributions include (a) curating a comprehensive corpus of semiconductor-related texts, (b) creating a foundational model with in-depth semiconductor knowledge, and (c) introducing a framework for integrating expert knowledge, thereby advancing the evaluation process of domain-specific AI models. Through fine-tuning a pre-trained LLM using our curated dataset, we have shown that SemiKong outperforms larger, general-purpose LLMs in various semiconductor manufacturing and design tasks. Our extensive experiments underscore the importance of developing domain-specific LLMs as a foundation for company- or tool-specific proprietary models, paving the way for further research and applications in the semiconductor domain. Code and dataset will be available at https://github.com/aitomatic/semikong
Enhancing Q&A with Domain-Specific Fine-Tuning and Iterative Reasoning: A Comparative Study
Apr 17, 2024



Abstract:This paper investigates the impact of domain-specific model fine-tuning and of reasoning mechanisms on the performance of question-answering (Q&A) systems powered by large language models (LLMs) and Retrieval-Augmented Generation (RAG). Using the FinanceBench SEC financial filings dataset, we observe that, for RAG, combining a fine-tuned embedding model with a fine-tuned LLM achieves better accuracy than generic models, with relatively greater gains attributable to fine-tuned embedding models. Additionally, employing reasoning iterations on top of RAG delivers an even bigger jump in performance, enabling the Q&A systems to get closer to human-expert quality. We discuss the implications of such findings, propose a structured technical design space capturing major technical components of Q&A AI, and provide recommendations for making high-impact technical choices for such components. We plan to follow up on this work with actionable guides for AI teams and further investigations into the impact of domain-specific augmentation in RAG and into agentic AI capabilities such as advanced planning and reasoning.
Multimodal Representation Learning of Cardiovascular Magnetic Resonance Imaging
Apr 16, 2023Abstract:Self-supervised learning is crucial for clinical imaging applications, given the lack of explicit labels in healthcare. However, conventional approaches that rely on precise vision-language alignment are not always feasible in complex clinical imaging modalities, such as cardiac magnetic resonance (CMR). CMR provides a comprehensive visualization of cardiac anatomy, physiology, and microstructure, making it challenging to interpret. Additionally, CMR reports require synthesizing information from sequences of images and different views, resulting in potentially weak alignment between the study and diagnosis report pair. To overcome these challenges, we propose \textbf{CMRformer}, a multimodal learning framework to jointly learn sequences of CMR images and associated cardiologist's reports. Moreover, one of the major obstacles to improving CMR study is the lack of large, publicly available datasets. To bridge this gap, we collected a large \textbf{CMR dataset}, which consists of 13,787 studies from clinical cases. By utilizing our proposed CMRformer and our collected dataset, we achieved remarkable performance in real-world clinical tasks, such as CMR image retrieval and diagnosis report retrieval. Furthermore, the learned representations are evaluated to be practically helpful for downstream applications, such as disease classification. Our work could potentially expedite progress in the CMR study and lead to more accurate and effective diagnosis and treatment.
A Multiagent CyberBattleSim for RL Cyber Operation Agents
Apr 03, 2023Abstract:Hardening cyber physical assets is both crucial and labor-intensive. Recently, Machine Learning (ML) in general and Reinforcement Learning RL) more specifically has shown great promise to automate tasks that otherwise would require significant human insight/intelligence. The development of autonomous RL agents requires a suitable training environment that allows us to quickly evaluate various alternatives, in particular how to arrange training scenarios that pit attackers and defenders against each other. CyberBattleSim is a training environment that supports the training of red agents, i.e., attackers. We added the capability to train blue agents, i.e., defenders. The paper describes our changes and reports on the results we obtained when training blue agents, either in isolation or jointly with red agents. Our results show that training a blue agent does lead to stronger defenses against attacks. In particular, training a blue agent jointly with a red agent increases the blue agent's capability to thwart sophisticated red agents.
Accelerated and Quantitative 3D Semisolid MT/CEST Imaging using a Generative Adversarial Network (GAN-CEST)
Jul 22, 2022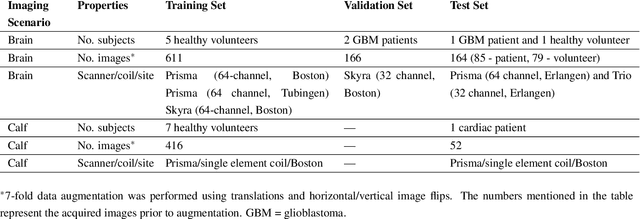
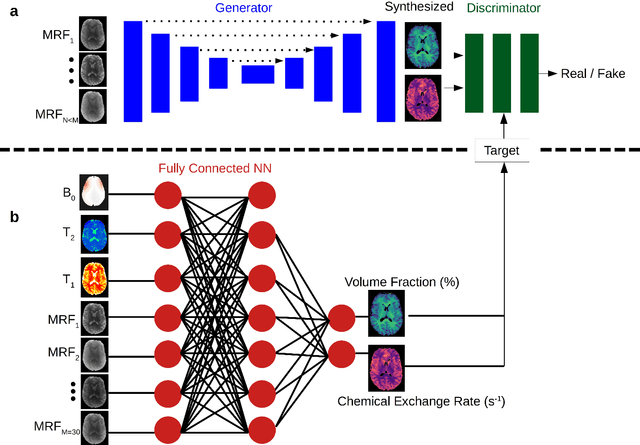
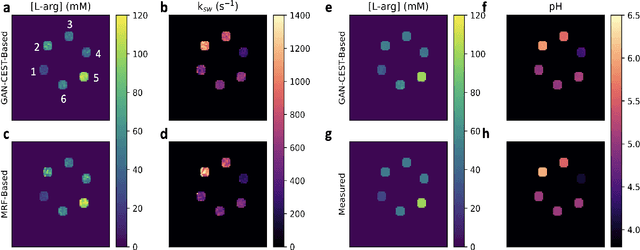
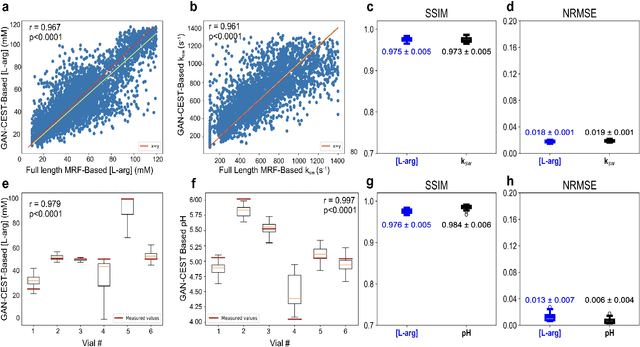
Abstract:Purpose: To substantially shorten the acquisition time required for quantitative 3D chemical exchange saturation transfer (CEST) and semisolid magnetization transfer (MT) imaging and allow for rapid chemical exchange parameter map reconstruction. Methods: Three-dimensional CEST and MT magnetic resonance fingerprinting (MRF) datasets of L-arginine phantoms, whole-brains, and calf muscles from healthy volunteers, cancer patients, and cardiac patients were acquired using 3T clinical scanners at 3 different sites, using 3 different scanner models and coils. A generative adversarial network supervised framework (GAN-CEST) was then designed and trained to learn the mapping from a reduced input data space to the quantitative exchange parameter space, while preserving perceptual and quantitative content. Results: The GAN-CEST 3D acquisition time was 42-52 seconds, 70% shorter than CEST-MRF. The quantitative reconstruction of the entire brain took 0.8 seconds. An excellent agreement was observed between the ground truth and GAN-based L-arginine concentration and pH values (Pearson's r > 0.97, NRMSE < 1.5%). GAN-CEST images from a brain-tumor subject yielded a semi-solid volume fraction and exchange rate NRMSE of 3.8$\pm$1.3% and 4.6$\pm$1.3%, respectively, and SSIM of 96.3$\pm$1.6% and 95.0$\pm$2.4%, respectively. The mapping of the calf-muscle exchange parameters in a cardiac patient, yielded NRMSE < 7% and SSIM > 94% for the semi-solid exchange parameters. In regions with large susceptibility artifacts, GAN-CEST has demonstrated improved performance and reduced noise compared to MRF. Conclusion: GAN-CEST can substantially reduce the acquisition time for quantitative semisolid MT/CEST mapping, while retaining performance even when facing pathologies and scanner models that were not available during training.
Interaction of a priori Anatomic Knowledge with Self-Supervised Contrastive Learning in Cardiac Magnetic Resonance Imaging
May 25, 2022



Abstract:Training deep learning models on cardiac magnetic resonance imaging (CMR) can be a challenge due to the small amount of expert generated labels and inherent complexity of data source. Self-supervised contrastive learning (SSCL) has recently been shown to boost performance in several medical imaging tasks. However, it is unclear how much the pre-trained representation reflects the primary organ of interest compared to spurious surrounding tissue. In this work, we evaluate the optimal method of incorporating prior knowledge of anatomy into a SSCL training paradigm. Specifically, we evaluate using a segmentation network to explicitly local the heart in CMR images, followed by SSCL pretraining in multiple diagnostic tasks. We find that using a priori knowledge of anatomy can greatly improve the downstream diagnostic performance. Furthermore, SSCL pre-training with in-domain data generally improved downstream performance and more human-like saliency compared to end-to-end training and ImageNet pre-trained networks. However, introducing anatomic knowledge to pre-training generally does not have significant impact.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge