Ramesh Basnet
Senior Member, IEEE
Interaction of a priori Anatomic Knowledge with Self-Supervised Contrastive Learning in Cardiac Magnetic Resonance Imaging
May 25, 2022



Abstract:Training deep learning models on cardiac magnetic resonance imaging (CMR) can be a challenge due to the small amount of expert generated labels and inherent complexity of data source. Self-supervised contrastive learning (SSCL) has recently been shown to boost performance in several medical imaging tasks. However, it is unclear how much the pre-trained representation reflects the primary organ of interest compared to spurious surrounding tissue. In this work, we evaluate the optimal method of incorporating prior knowledge of anatomy into a SSCL training paradigm. Specifically, we evaluate using a segmentation network to explicitly local the heart in CMR images, followed by SSCL pretraining in multiple diagnostic tasks. We find that using a priori knowledge of anatomy can greatly improve the downstream diagnostic performance. Furthermore, SSCL pre-training with in-domain data generally improved downstream performance and more human-like saliency compared to end-to-end training and ImageNet pre-trained networks. However, introducing anatomic knowledge to pre-training generally does not have significant impact.
Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge
Jul 11, 2020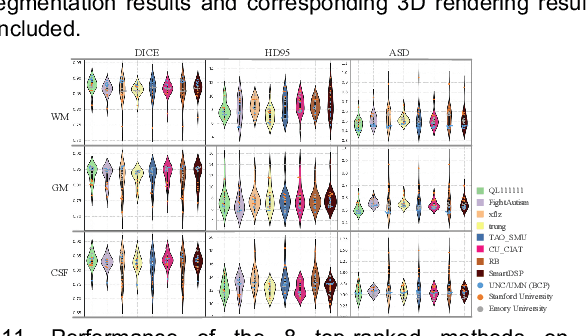
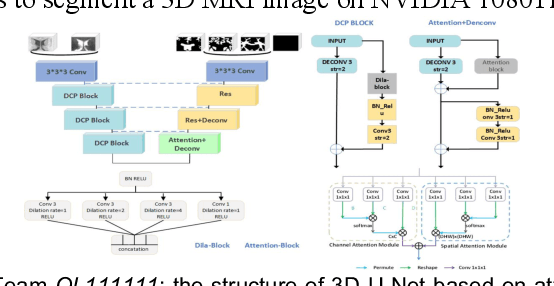
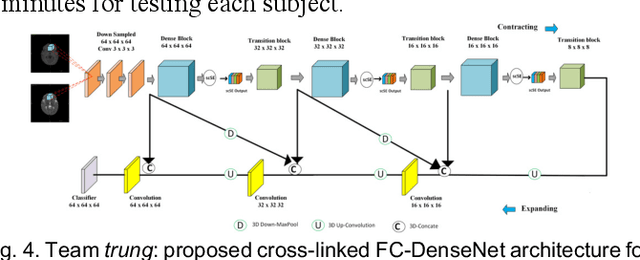
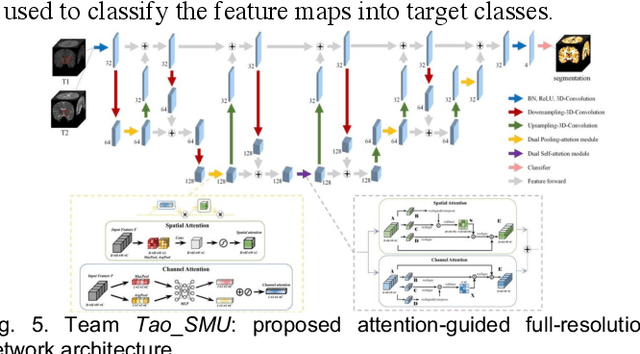
Abstract:To better understand early brain growth patterns in health and disorder, it is critical to accurately segment infant brain magnetic resonance (MR) images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Deep learning-based methods have achieved state-of-the-art performance; however, one of major limitations is that the learning-based methods may suffer from the multi-site issue, that is, the models trained on a dataset from one site may not be applicable to the datasets acquired from other sites with different imaging protocols/scanners. To promote methodological development in the community, iSeg-2019 challenge (http://iseg2019.web.unc.edu) provides a set of 6-month infant subjects from multiple sites with different protocols/scanners for the participating methods. Training/validation subjects are from UNC (MAP) and testing subjects are from UNC/UMN (BCP), Stanford University, and Emory University. By the time of writing, there are 30 automatic segmentation methods participating in iSeg-2019. We review the 8 top-ranked teams by detailing their pipelines/implementations, presenting experimental results and evaluating performance in terms of the whole brain, regions of interest, and gyral landmark curves. We also discuss their limitations and possible future directions for the multi-site issue. We hope that the multi-site dataset in iSeg-2019 and this review article will attract more researchers on the multi-site issue.
Statistical Selection of CNN-Based Audiovisual Features for Instantaneous Estimation of Human Emotional States
Aug 23, 2017
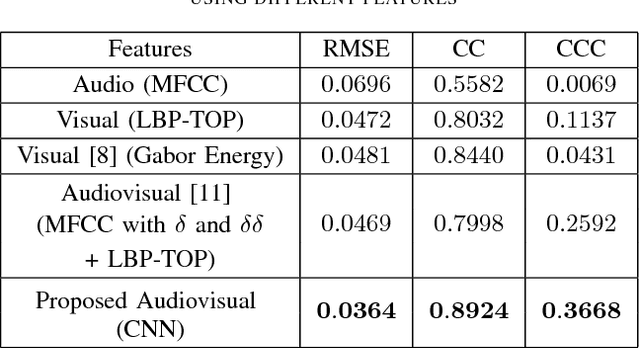
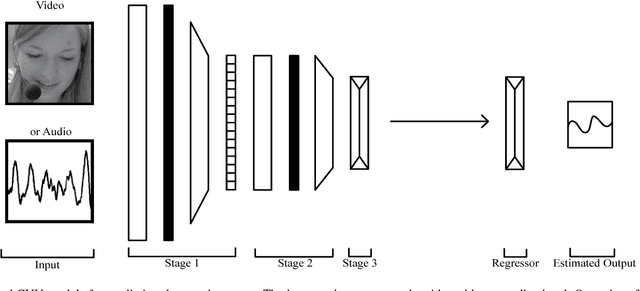
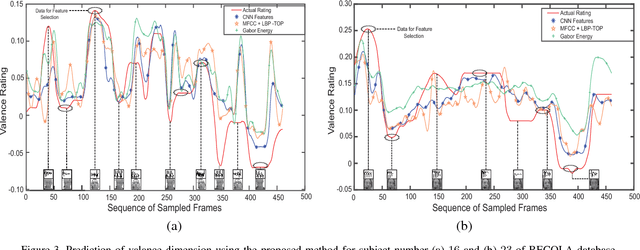
Abstract:Automatic prediction of continuous-level emotional state requires selection of suitable affective features to develop a regression system based on supervised machine learning. This paper investigates the performance of features statistically learned using convolutional neural networks for instantaneously predicting the continuous dimensions of emotional states. Features with minimum redundancy and maximum relevancy are chosen by using the mutual information-based selection process. The performance of frame-by-frame prediction of emotional state using the moderate length features as proposed in this paper is evaluated on spontaneous and naturalistic human-human conversation of RECOLA database. Experimental results show that the proposed model can be used for instantaneous prediction of emotional state with an accuracy higher than traditional audio or video features that are used for affective computation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge