Makiya Nakashima
Multimodal Representation Learning of Cardiovascular Magnetic Resonance Imaging
Apr 16, 2023Abstract:Self-supervised learning is crucial for clinical imaging applications, given the lack of explicit labels in healthcare. However, conventional approaches that rely on precise vision-language alignment are not always feasible in complex clinical imaging modalities, such as cardiac magnetic resonance (CMR). CMR provides a comprehensive visualization of cardiac anatomy, physiology, and microstructure, making it challenging to interpret. Additionally, CMR reports require synthesizing information from sequences of images and different views, resulting in potentially weak alignment between the study and diagnosis report pair. To overcome these challenges, we propose \textbf{CMRformer}, a multimodal learning framework to jointly learn sequences of CMR images and associated cardiologist's reports. Moreover, one of the major obstacles to improving CMR study is the lack of large, publicly available datasets. To bridge this gap, we collected a large \textbf{CMR dataset}, which consists of 13,787 studies from clinical cases. By utilizing our proposed CMRformer and our collected dataset, we achieved remarkable performance in real-world clinical tasks, such as CMR image retrieval and diagnosis report retrieval. Furthermore, the learned representations are evaluated to be practically helpful for downstream applications, such as disease classification. Our work could potentially expedite progress in the CMR study and lead to more accurate and effective diagnosis and treatment.
Interaction of a priori Anatomic Knowledge with Self-Supervised Contrastive Learning in Cardiac Magnetic Resonance Imaging
May 25, 2022



Abstract:Training deep learning models on cardiac magnetic resonance imaging (CMR) can be a challenge due to the small amount of expert generated labels and inherent complexity of data source. Self-supervised contrastive learning (SSCL) has recently been shown to boost performance in several medical imaging tasks. However, it is unclear how much the pre-trained representation reflects the primary organ of interest compared to spurious surrounding tissue. In this work, we evaluate the optimal method of incorporating prior knowledge of anatomy into a SSCL training paradigm. Specifically, we evaluate using a segmentation network to explicitly local the heart in CMR images, followed by SSCL pretraining in multiple diagnostic tasks. We find that using a priori knowledge of anatomy can greatly improve the downstream diagnostic performance. Furthermore, SSCL pre-training with in-domain data generally improved downstream performance and more human-like saliency compared to end-to-end training and ImageNet pre-trained networks. However, introducing anatomic knowledge to pre-training generally does not have significant impact.
An Ensemble Approach toward Automated Variable Selection for Network Anomaly Detection
Oct 28, 2019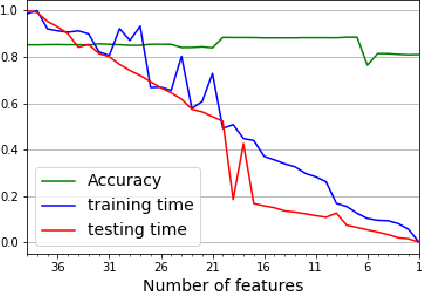
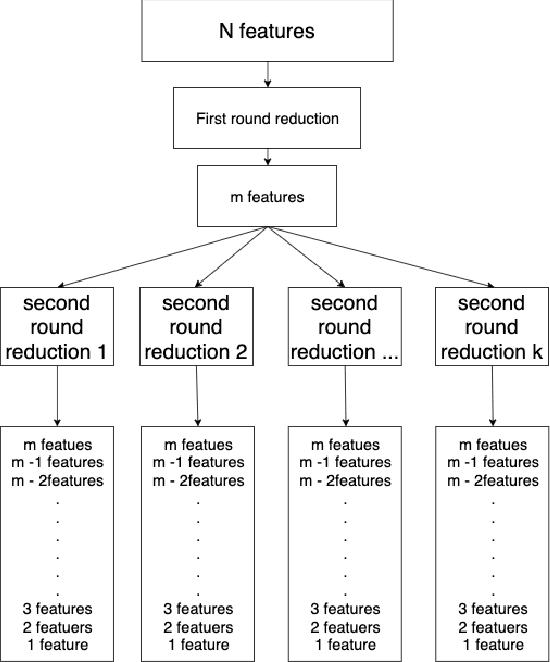
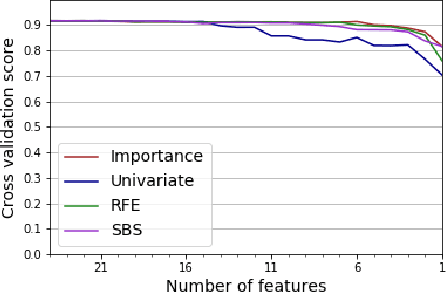
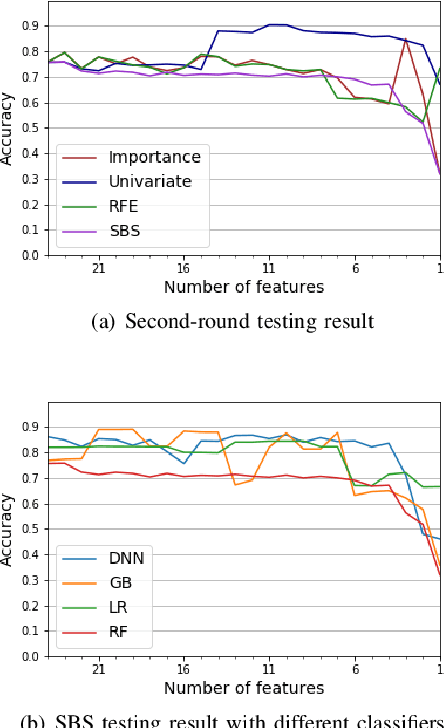
Abstract:While variable selection is essential to optimize the learning complexity by prioritizing features, automating the selection process is preferred since it requires laborious efforts with intensive analysis otherwise. However, it is not an easy task to enable the automation due to several reasons. First, selection techniques often need a condition to terminate the reduction process, for example, by using a threshold or the number of features to stop, and searching an adequate stopping condition is highly challenging. Second, it is uncertain that the reduced variable set would work well; our preliminary experimental result shows that well-known selection techniques produce different sets of variables as a result of reduction (even with the same termination condition), and it is hard to estimate which of them would work the best in future testing. In this paper, we demonstrate the potential power of our approach to the automation of selection process that incorporates well-known selection methods identifying important variables. Our experimental results with two public network traffic data (UNSW-NB15 and IDS2017) show that our proposed method identifies a small number of core variables, with which it is possible to approximate the performance to the one with the entire variables.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge