Nikita Vladimirov
Multi-Parameter Molecular MRI Quantification using Physics-Informed Self-Supervised Learning
Nov 10, 2024Abstract:Biophysical model fitting plays a key role in obtaining quantitative parameters from physiological signals and images. However, the model complexity for molecular magnetic resonance imaging (MRI) often translates into excessive computation time, which makes clinical use impractical. Here, we present a generic computational approach for solving the parameter extraction inverse problem posed by ordinary differential equation (ODE) modeling coupled with experimental measurement of the system dynamics. This is achieved by formulating a numerical ODE solver to function as a step-wise analytical one, thereby making it compatible with automatic differentiation-based optimization. This enables efficient gradient-based model fitting, and provides a new approach to parameter quantification based on self-supervised learning from a single data observation. The neural-network-based train-by-fit pipeline was used to quantify semisolid magnetization transfer (MT) and chemical exchange saturation transfer (CEST) amide proton exchange parameters in the human brain, in an in-vivo molecular MRI study (n=4). The entire pipeline of the first whole brain quantification was completed in 18.3$\pm$8.3 minutes, which is an order-of-magnitude faster than comparable alternatives. Reusing the single-subject-trained network for inference in new subjects took 1.0$\pm$0.2 s, to provide results in agreement with literature values and scan-specific fit results (Pearson's r>0.98, p<0.0001).
Dynamic and Rapid Deep Synthesis of Molecular MRI Signals
May 30, 2023Abstract:Model-driven analysis of biophysical phenomena is gaining increased attention and utility for medical imaging applications. In magnetic resonance imaging (MRI), the availability of well-established models for describing the relations between the nuclear magnetization, tissue properties, and the externally applied magnetic fields has enabled the prediction of image contrast and served as a powerful tool for designing the imaging protocols that are now routinely used in the clinic. Recently, various advanced imaging techniques have relied on these models for image reconstruction, quantitative tissue parameter extraction, and automatic optimization of acquisition protocols. In molecular MRI, however, the increased complexity of the imaging scenario, where the signals from various chemical compounds and multiple proton pools must be accounted for, results in exceedingly long model simulation times, severely hindering the progress of this approach and its dissemination for various clinical applications. Here, we show that a deep-learning-based system can capture the nonlinear relations embedded in the molecular MRI Bloch-McConnell model, enabling a rapid and accurate generation of biologically realistic synthetic data. The applicability of this simulated data for in-silico, in-vitro, and in-vivo imaging applications is then demonstrated for chemical exchange saturation transfer (CEST) and semisolid macromolecule magnetization transfer (MT) analysis and quantification. The proposed approach yielded 78%-99% acceleration in data synthesis time while retaining excellent agreement with the ground truth (Pearson's r$>$0.99, p$<$0.0001, normalized root mean square error $<$3%).
CNN-based fully automatic wrist cartilage volume quantification in MR Image
Jun 22, 2022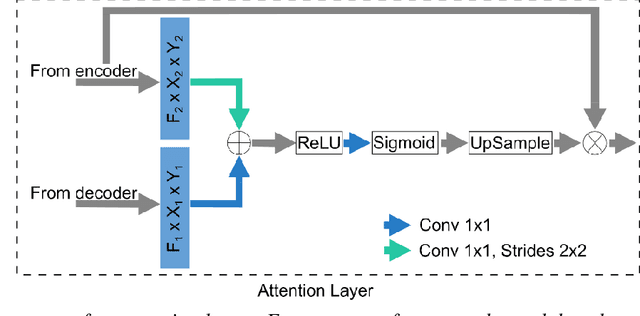
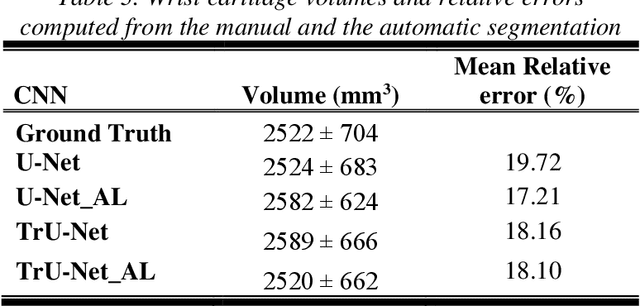
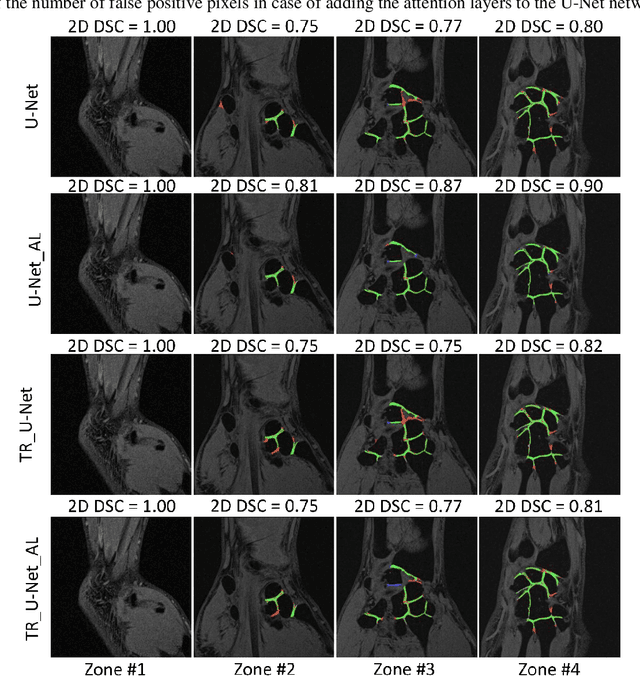
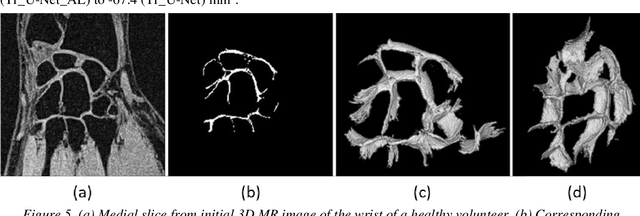
Abstract:Detection of cartilage loss is crucial for the diagnosis of osteo- and rheumatoid arthritis. A large number of automatic segmentation tools have been reported so far for cartilage assessment in magnetic resonance images of large joints. As compared to knee or hip, wrist cartilage has a more complex structure so that automatic tools developed for large joints are not expected to be operational for wrist cartilage segmentation. In that respect, a fully automatic wrist cartilage segmentation method would be of high clinical interest. We assessed the performance of four optimized variants of the U-Net architecture with truncation of its depth and addition of attention layers (U-Net_AL). The corresponding results were compared to those from a patch-based convolutional neural network (CNN) we previously designed. The segmentation quality was assessed on the basis of a comparative analysis with manual segmentation using several morphological (2D DSC, 3D DSC, precision) and a volumetric metrics. The four networks outperformed the patch-based CNN in terms of segmentation homogeneity and quality. The median 3D DSC value computed with the U-Net_AL (0.817) was significantly larger than the corresponding DSC values computed with the other networks. In addition, the U-Net_AL CNN provided the lowest mean volume error (17%) and the highest Pearson correlation coefficient (0.765) with respect to the ground truth. Of interest, the reproducibility computed from using U-Net_AL was larger than the reproducibility of the manual segmentation. U-net convolutional neural network with additional attention layers provides the best wrist cartilage segmentation performance. In order to be used in clinical conditions, the trained network can be fine-tuned on a dataset representing a group of specific patients. The error of cartilage volume measurement should be assessed independently using a non-MRI method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge