Emmanouil Nikolakakis
Language Models for Music Medicine Generation
Nov 13, 2024
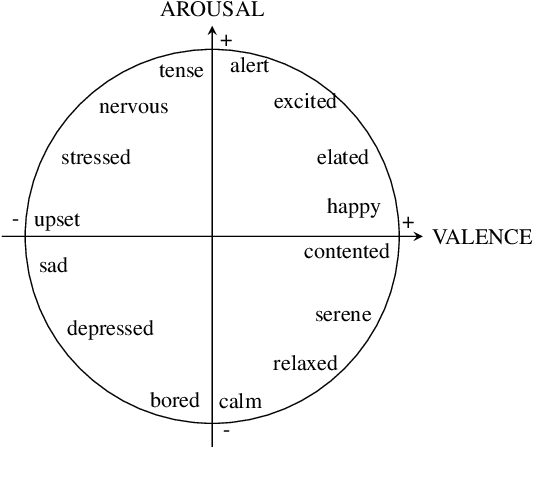
Abstract:Music therapy has been shown in recent years to provide multiple health benefits related to emotional wellness. In turn, maintaining a healthy emotional state has proven to be effective for patients undergoing treatment, such as Parkinson's patients or patients suffering from stress and anxiety. We propose fine-tuning MusicGen, a music-generating transformer model, to create short musical clips that assist patients in transitioning from negative to desired emotional states. Using low-rank decomposition fine-tuning on the MTG-Jamendo Dataset with emotion tags, we generate 30-second clips that adhere to the iso principle, guiding patients through intermediate states in the valence-arousal circumplex. The generated music is evaluated using a music emotion recognition model to ensure alignment with intended emotions. By concatenating these clips, we produce a 15-minute "music medicine" resembling a music therapy session. Our approach is the first model to leverage Language Models to generate music medicine. Ultimately, the output is intended to be used as a temporary relief between music therapy sessions with a board-certified therapist.
AFEN: Respiratory Disease Classification using Ensemble Learning
May 08, 2024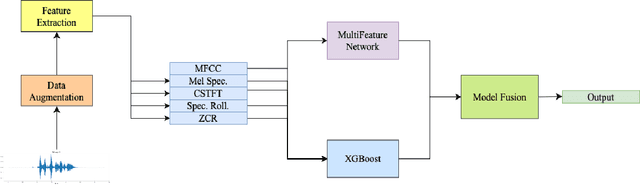
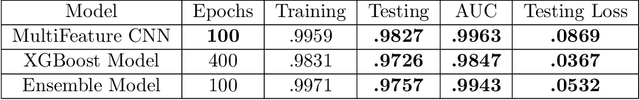
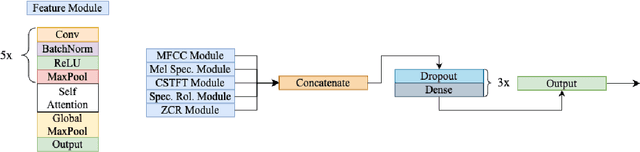
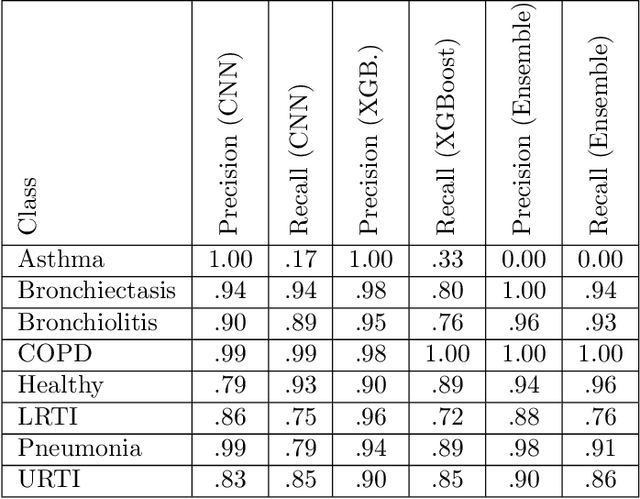
Abstract:We present AFEN (Audio Feature Ensemble Learning), a model that leverages Convolutional Neural Networks (CNN) and XGBoost in an ensemble learning fashion to perform state-of-the-art audio classification for a range of respiratory diseases. We use a meticulously selected mix of audio features which provide the salient attributes of the data and allow for accurate classification. The extracted features are then used as an input to two separate model classifiers 1) a multi-feature CNN classifier and 2) an XGBoost Classifier. The outputs of the two models are then fused with the use of soft voting. Thus, by exploiting ensemble learning, we achieve increased robustness and accuracy. We evaluate the performance of the model on a database of 920 respiratory sounds, which undergoes data augmentation techniques to increase the diversity of the data and generalizability of the model. We empirically verify that AFEN sets a new state-of-the-art using Precision and Recall as metrics, while decreasing training time by 60%.
BMapOpt: Optimization of Brain Tissue Probability Maps using a Differentiable MRI Simulator
Apr 23, 2024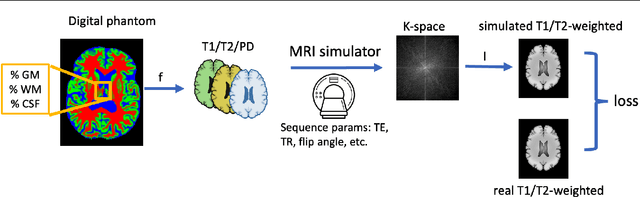
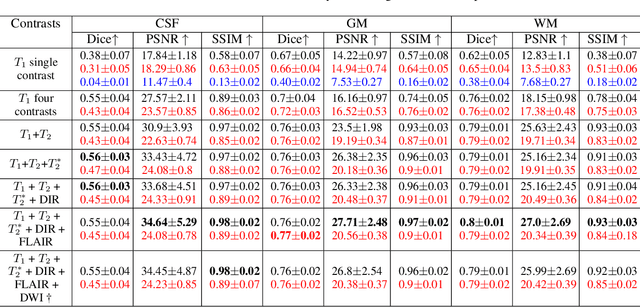
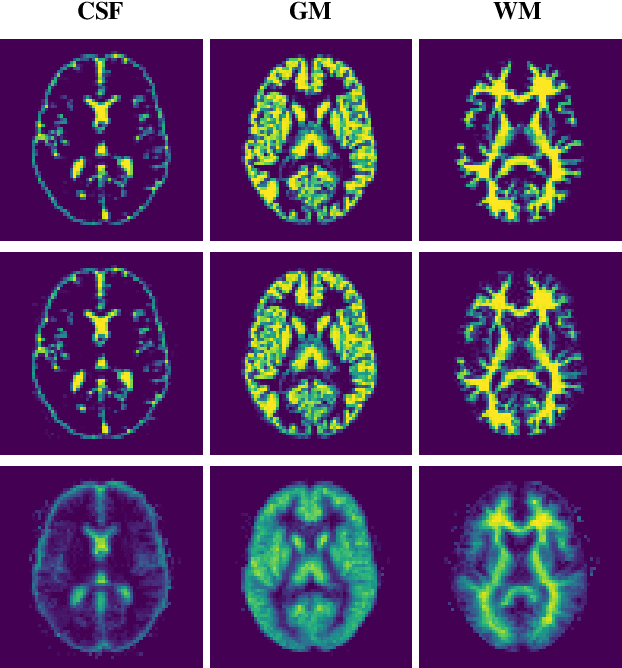
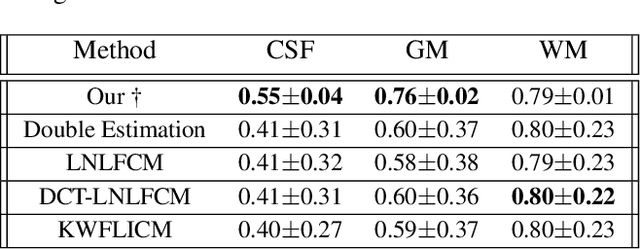
Abstract:Reconstructing digital brain phantoms in the form of multi-channeled brain tissue probability maps for individual subjects is essential for capturing brain anatomical variability, understanding neurological diseases, as well as for testing image processing methods. We demonstrate the first framework that optimizes brain tissue probability maps (Gray Matter - GM, White Matter - WM, and Cerebrospinal fluid - CSF) with the help of a Physics-based differentiable MRI simulator that models the magnetization signal at each voxel in the image. Given an observed $T_1$/$T_2$-weighted MRI scan, the corresponding clinical MRI sequence, and the MRI differentiable simulator, we optimize the simulator's input probability maps by back-propagating the L2 loss between the simulator's output and the $T_1$/$T_2$-weighted scan. This approach has the significant advantage of not relying on any training data, and instead uses the strong inductive bias of the MRI simulator. We tested the model on 20 scans from the BrainWeb database and demonstrate a highly accurate reconstruction of GM, WM, and CSF.
GaSpCT: Gaussian Splatting for Novel CT Projection View Synthesis
Apr 04, 2024



Abstract:We present GaSpCT, a novel view synthesis and 3D scene representation method used to generate novel projection views for Computer Tomography (CT) scans. We adapt the Gaussian Splatting framework to enable novel view synthesis in CT based on limited sets of 2D image projections and without the need for Structure from Motion (SfM) methodologies. Therefore, we reduce the total scanning duration and the amount of radiation dose the patient receives during the scan. We adapted the loss function to our use-case by encouraging a stronger background and foreground distinction using two sparsity promoting regularizers: a beta loss and a total variation (TV) loss. Finally, we initialize the Gaussian locations across the 3D space using a uniform prior distribution of where the brain's positioning would be expected to be within the field of view. We evaluate the performance of our model using brain CT scans from the Parkinson's Progression Markers Initiative (PPMI) dataset and demonstrate that the rendered novel views closely match the original projection views of the simulated scan, and have better performance than other implicit 3D scene representations methodologies. Furthermore, we empirically observe reduced training time compared to neural network based image synthesis for sparse-view CT image reconstruction. Finally, the memory requirements of the Gaussian Splatting representations are reduced by 17% compared to the equivalent voxel grid image representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge