Manuel Schmidt
Comprehensive Multimodal Deep Learning Survival Prediction Enabled by a Transformer Architecture: A Multicenter Study in Glioblastoma
May 21, 2024Abstract:Background: This research aims to improve glioblastoma survival prediction by integrating MR images, clinical and molecular-pathologic data in a transformer-based deep learning model, addressing data heterogeneity and performance generalizability. Method: We propose and evaluate a transformer-based non-linear and non-proportional survival prediction model. The model employs self-supervised learning techniques to effectively encode the high-dimensional MRI input for integration with non-imaging data using cross-attention. To demonstrate model generalizability, the model is assessed with the time-dependent concordance index (Cdt) in two training setups using three independent public test sets: UPenn-GBM, UCSF-PDGM, and RHUH-GBM, each comprising 378, 366, and 36 cases, respectively. Results: The proposed transformer model achieved promising performance for imaging as well as non-imaging data, effectively integrating both modalities for enhanced performance (UPenn-GBM test-set, imaging Cdt 0.645, multimodal Cdt 0.707) while outperforming state-of-the-art late-fusion 3D-CNN-based models. Consistent performance was observed across the three independent multicenter test sets with Cdt values of 0.707 (UPenn-GBM, internal test set), 0.672 (UCSF-PDGM, first external test set) and 0.618 (RHUH-GBM, second external test set). The model achieved significant discrimination between patients with favorable and unfavorable survival for all three datasets (logrank p 1.9\times{10}^{-8}, 9.7\times{10}^{-3}, and 1.2\times{10}^{-2}). Conclusions: The proposed transformer-based survival prediction model integrates complementary information from diverse input modalities, contributing to improved glioblastoma survival prediction compared to state-of-the-art methods. Consistent performance was observed across institutions supporting model generalizability.
Multicenter Privacy-Preserving Model Training for Deep Learning Brain Metastases Autosegmentation
May 17, 2024



Abstract:Objectives: This work aims to explore the impact of multicenter data heterogeneity on deep learning brain metastases (BM) autosegmentation performance, and assess the efficacy of an incremental transfer learning technique, namely learning without forgetting (LWF), to improve model generalizability without sharing raw data. Materials and methods: A total of six BM datasets from University Hospital Erlangen (UKER), University Hospital Zurich (USZ), Stanford, UCSF, NYU and BraTS Challenge 2023 on BM segmentation were used for this evaluation. First, the multicenter performance of a convolutional neural network (DeepMedic) for BM autosegmentation was established for exclusive single-center training and for training on pooled data, respectively. Subsequently bilateral collaboration was evaluated, where a UKER pretrained model is shared to another center for further training using transfer learning (TL) either with or without LWF. Results: For single-center training, average F1 scores of BM detection range from 0.625 (NYU) to 0.876 (UKER) on respective single-center test data. Mixed multicenter training notably improves F1 scores at Stanford and NYU, with negligible improvement at other centers. When the UKER pretrained model is applied to USZ, LWF achieves a higher average F1 score (0.839) than naive TL (0.570) and single-center training (0.688) on combined UKER and USZ test data. Naive TL improves sensitivity and contouring accuracy, but compromises precision. Conversely, LWF demonstrates commendable sensitivity, precision and contouring accuracy. When applied to Stanford, similar performance was observed. Conclusion: Data heterogeneity results in varying performance in BM autosegmentation, posing challenges to model generalizability. LWF is a promising approach to peer-to-peer privacy-preserving model training.
Risk Classification of Brain Metastases via Radiomics, Delta-Radiomics and Machine Learning
Feb 17, 2023



Abstract:Stereotactic radiotherapy (SRT) is one of the most important treatment for patients with brain metastases (BM). Conventionally, following SRT patients are monitored by serial imaging and receive salvage treatments in case of significant tumor growth. We hypothesized that using radiomics and machine learning (ML), metastases at high risk for subsequent progression could be identified during follow-up prior to the onset of significant tumor growth, enabling personalized follow-up intervals and early selection for salvage treatment. All experiments are performed on a dataset from clinical routine of the Radiation Oncology department of the University Hospital Erlangen (UKER). The classification is realized via the maximum-relevance minimal-redundancy (MRMR) technique and support vector machines (SVM). The pipeline leads to a classification with a mean area under the curve (AUC) score of 0.83 in internal cross-validation and allows a division of the cohort into two subcohorts that differ significantly in their median time to progression (low-risk metastasis (LRM): 17.3 months, high-risk metastasis (HRM): 9.6 months, p < 0.01). The classification performance is especially enhanced by the analysis of medical images from different points in time (AUC 0.53 -> AUC 0.74). The results indicate that risk stratification of BM based on radiomics and machine learning during post-SRT follow-up is possible with good accuracy and should be further pursued to personalize and improve post-SRT follow-up.
snapshot CEST++ : the next snapshot CEST for fast whole-brain APTw imaging at 3T
Jul 01, 2022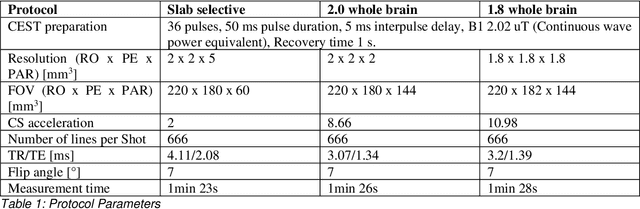
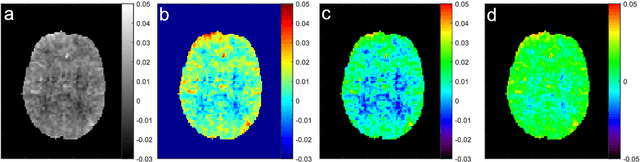
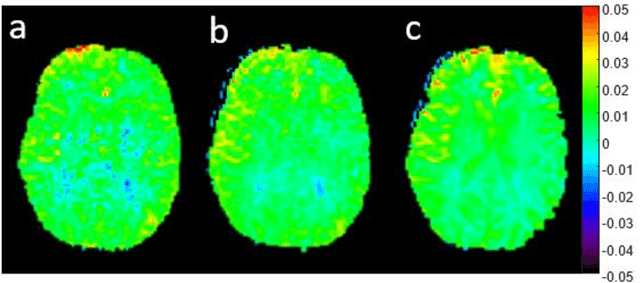
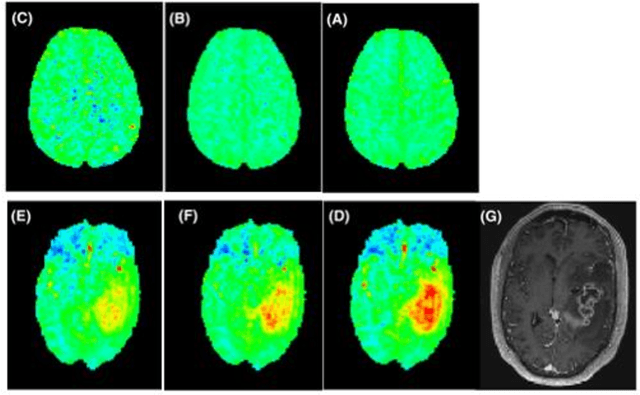
Abstract:CEST suffers from two main problems long acquisitin times or restricted coverage as well as incoherent protocol settings. In this paper we give suggestions on how to optimise your protocol settings fro CEST and present one setting for APT CEST. To increase the coverage while keeping the acquisition time constant we suggest using a spatial temporal Compressed Sensing approach. Finally, 1.8mm isotropic whole brain APT CEST maps can be acquired in a little bit less than 2min with a fully integrated online reconstruction. This will pave the way to an even further clinical use of CEST.
Continual Learning for Peer-to-Peer Federated Learning: A Study on Automated Brain Metastasis Identification
Apr 30, 2022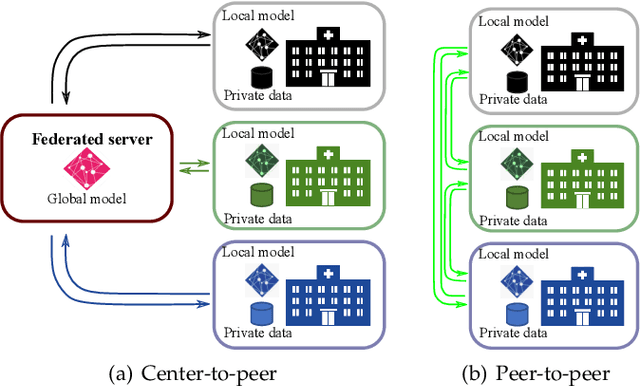
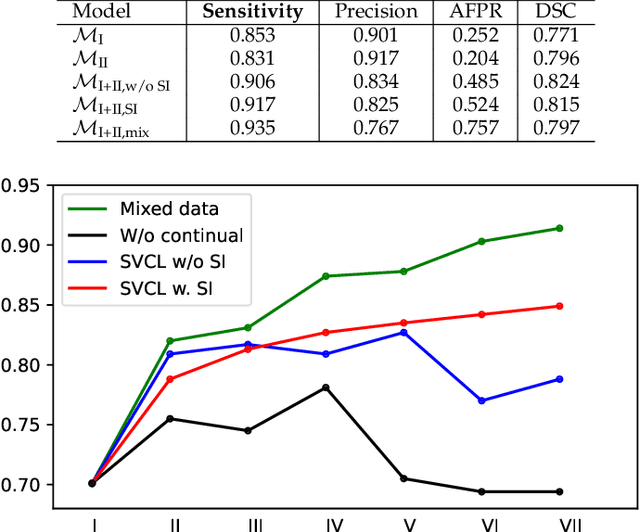
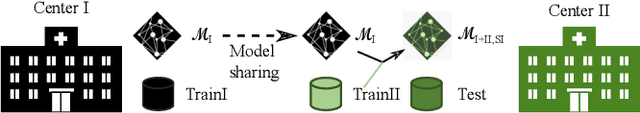
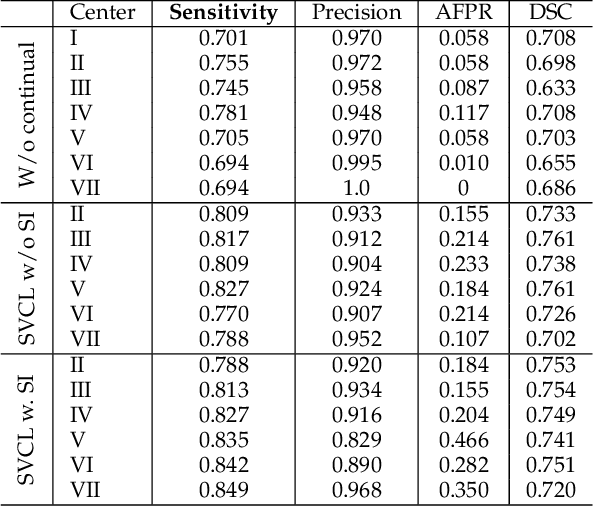
Abstract:Due to data privacy constraints, data sharing among multiple centers is restricted. Continual learning, as one approach to peer-to-peer federated learning, can promote multicenter collaboration on deep learning algorithm development by sharing intermediate models instead of training data. This work aims to investigate the feasibility of continual learning for multicenter collaboration on an exemplary application of brain metastasis identification using DeepMedic. 920 T1 MRI contrast enhanced volumes are split to simulate multicenter collaboration scenarios. A continual learning algorithm, synaptic intelligence (SI), is applied to preserve important model weights for training one center after another. In a bilateral collaboration scenario, continual learning with SI achieves a sensitivity of 0.917, and naive continual learning without SI achieves a sensitivity of 0.906, while two models trained on internal data solely without continual learning achieve sensitivity of 0.853 and 0.831 only. In a seven-center multilateral collaboration scenario, the models trained on internal datasets (100 volumes each center) without continual learning obtain a mean sensitivity value of 0.699. With single-visit continual learning (i.e., the shared model visits each center only once during training), the sensitivity is improved to 0.788 and 0.849 without SI and with SI, respectively. With iterative continual learning (i.e., the shared model revisits each center multiple times during training), the sensitivity is further improved to 0.914, which is identical to the sensitivity using mixed data for training. Our experiments demonstrate that continual learning can improve brain metastasis identification performance for centers with limited data. This study demonstrates the feasibility of applying continual learning for peer-to-peer federated learning in multicenter collaboration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge