Manuel A. Schmidt
A Self-supervised Multimodal Deep Learning Approach to Differentiate Post-radiotherapy Progression from Pseudoprogression in Glioblastoma
Feb 06, 2025



Abstract:Accurate differentiation of pseudoprogression (PsP) from True Progression (TP) following radiotherapy (RT) in glioblastoma (GBM) patients is crucial for optimal treatment planning. However, this task remains challenging due to the overlapping imaging characteristics of PsP and TP. This study therefore proposes a multimodal deep-learning approach utilizing complementary information from routine anatomical MR images, clinical parameters, and RT treatment planning information for improved predictive accuracy. The approach utilizes a self-supervised Vision Transformer (ViT) to encode multi-sequence MR brain volumes to effectively capture both global and local context from the high dimensional input. The encoder is trained in a self-supervised upstream task on unlabeled glioma MRI datasets from the open BraTS2021, UPenn-GBM, and UCSF-PDGM datasets to generate compact, clinically relevant representations from FLAIR and T1 post-contrast sequences. These encoded MR inputs are then integrated with clinical data and RT treatment planning information through guided cross-modal attention, improving progression classification accuracy. This work was developed using two datasets from different centers: the Burdenko Glioblastoma Progression Dataset (n = 59) for training and validation, and the GlioCMV progression dataset from the University Hospital Erlangen (UKER) (n = 20) for testing. The proposed method achieved an AUC of 75.3%, outperforming the current state-of-the-art data-driven approaches. Importantly, the proposed approach relies on readily available anatomical MRI sequences, clinical data, and RT treatment planning information, enhancing its clinical feasibility. The proposed approach addresses the challenge of limited data availability for PsP and TP differentiation and could allow for improved clinical decision-making and optimized treatment plans for GBM patients.
Deep learning for brain metastasis detection and segmentation in longitudinal MRI data
Dec 28, 2021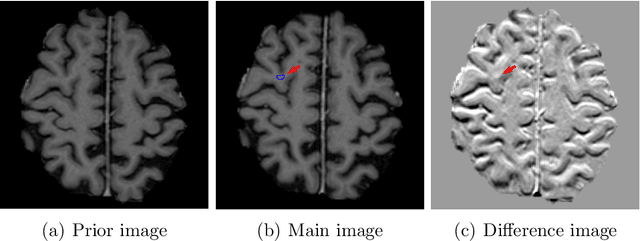
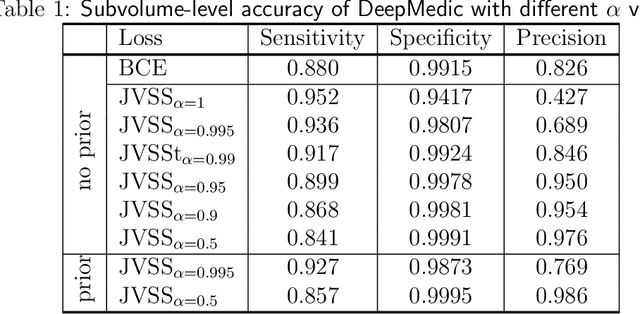
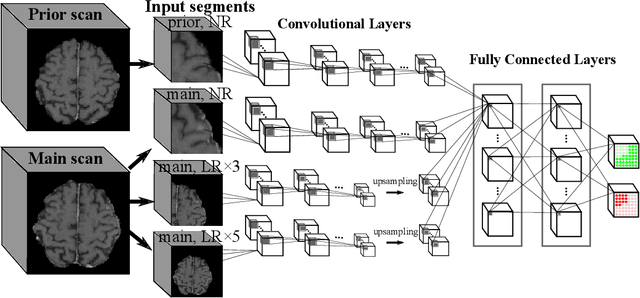
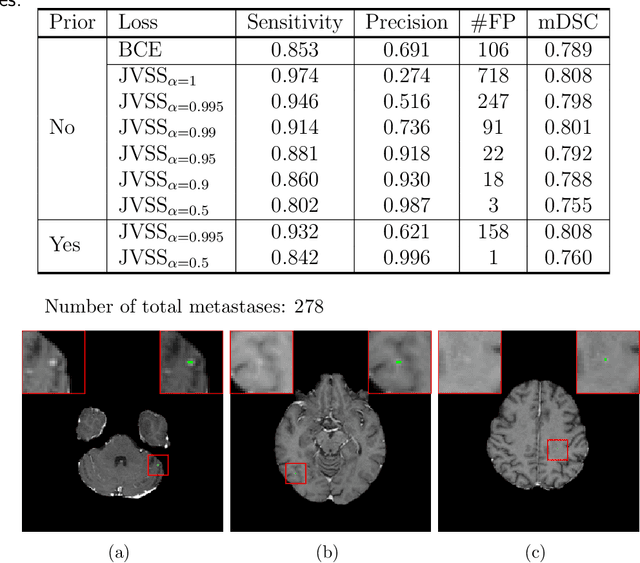
Abstract:Brain metastases occur frequently in patients with metastatic cancer. Early and accurate detection of brain metastases is very essential for treatment planning and prognosis in radiation therapy. To improve brain metastasis detection performance with deep learning, a custom detection loss called volume-level sensitivity-specificity (VSS) is proposed, which rates individual metastasis detection sensitivity and specificity in (sub-)volume levels. As sensitivity and precision are always a trade-off in a metastasis level, either a high sensitivity or a high precision can be achieved by adjusting the weights in the VSS loss without decline in dice score coefficient for segmented metastases. To reduce metastasis-like structures being detected as false positive metastases, a temporal prior volume is proposed as an additional input of the neural network. Our proposed VSS loss improves the sensitivity of brain metastasis detection, increasing the sensitivity from 86.7% to 95.5%. Alternatively, it improves the precision from 68.8% to 97.8%. With the additional temporal prior volume, about 45% of the false positive metastases are reduced in the high sensitivity model and the precision reaches 99.6% for the high specificity model. The mean dice coefficient for all metastases is about 0.81. With the ensemble of the high sensitivity and high specificity models, on average only 1.5 false positive metastases per patient needs further check, while the majority of true positive metastases are confirmed. The ensemble learning is able to distinguish high confidence true positive metastases from metastases candidates that require special expert review or further follow-up, being particularly well-fit to the requirements of expert support in real clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge