Marc Kachelrieß
Two-View Topogram-Based Anatomy-Guided CT Reconstruction for Prospective Risk Minimization
Jan 23, 2024Abstract:To facilitate a prospective estimation of CT effective dose and risk minimization process, a prospective spatial dose estimation and the known anatomical structures are expected. To this end, a CT reconstruction method is required to reconstruct CT volumes from as few projections as possible, i.e. by using the topograms, with anatomical structures as correct as possible. In this work, an optimized CT reconstruction model based on a generative adversarial network (GAN) is proposed. The GAN is trained to reconstruct 3D volumes from an anterior-posterior and a lateral CT projection. To enhance anatomical structures, a pre-trained organ segmentation network and the 3D perceptual loss are applied during the training phase, so that the model can then generate both organ-enhanced CT volume and the organ segmentation mask. The proposed method can reconstruct CT volumes with PSNR of 26.49, RMSE of 196.17, and SSIM of 0.64, compared to 26.21, 201.55 and 0.63 using the baseline method. In terms of the anatomical structure, the proposed method effectively enhances the organ shape and boundary and allows for a straight-forward identification of the relevant anatomical structures. We note that conventional reconstruction metrics fail to indicate the enhancement of anatomical structures. In addition to such metrics, the evaluation is expanded with assessing the organ segmentation performance. The average organ dice of the proposed method is 0.71 compared with 0.63 in baseline model, indicating the enhancement of anatomical structures.
Robustness Investigation on Deep Learning CT Reconstruction for Real-Time Dose Optimization
Dec 07, 2020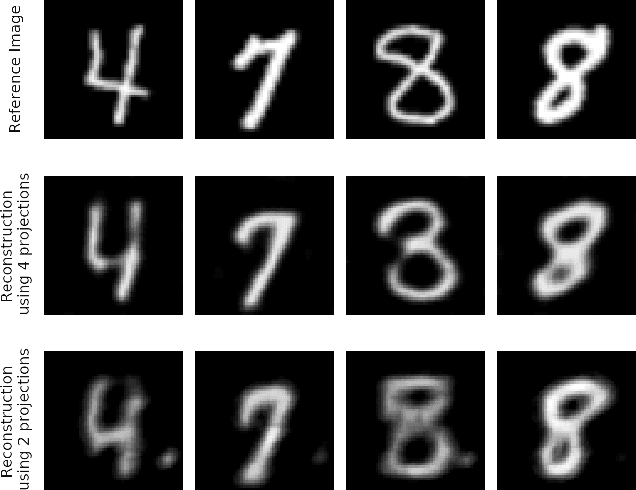


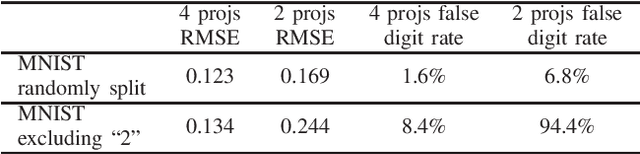
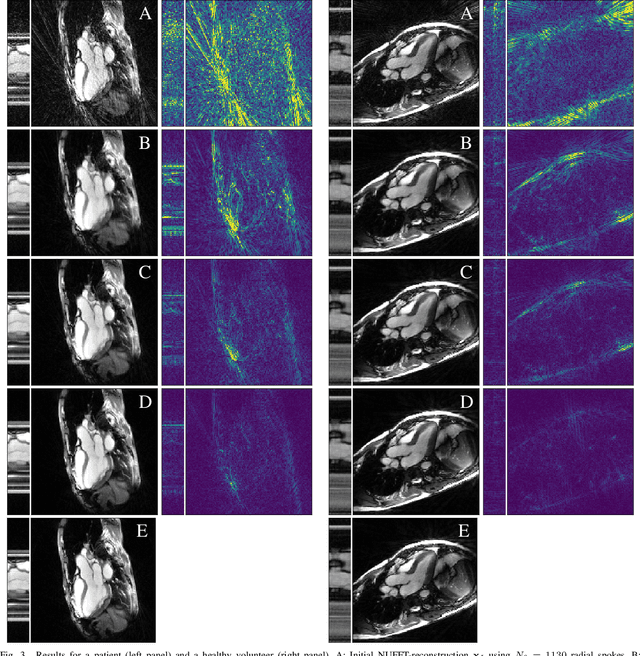
Abstract:In computed tomography (CT), automatic exposure control (AEC) is frequently used to reduce radiation dose exposure to patients. For organ-specific AEC, a preliminary CT reconstruction is necessary to estimate organ shapes for dose optimization, where only a few projections are allowed for real-time reconstruction. In this work, we investigate the performance of automated transform by manifold approximation (AUTOMAP) in such applications. For proof of concept, we investigate its performance on the MNIST dataset first, where the dataset containing all the 10 digits are randomly split into a training set and a test set. We train the AUTOMAP model for image reconstruction from 2 projections or 4 projections directly. The test results demonstrate that AUTOMAP is able to reconstruct most digits well with a false rate of 1.6% and 6.8% respectively. In our subsequent experiment, the MNIST dataset is split in a way that the training set contains 9 digits only while the test set contains the excluded digit only, for instance "2". In the test results, the digit "2"s are falsely predicted as "3" or "5" when using 2 projections for reconstruction, reaching a false rate of 94.4%. For the application in medical images, AUTOMAP is also trained on patients' CT images. The test images reach an average root-mean-square error of 290 HU. Although the coarse body outlines are well reconstructed, some organs are misshaped.
Neural Networks-based Regularization for Large-Scale Medical Image Reconstruction
Jan 22, 2020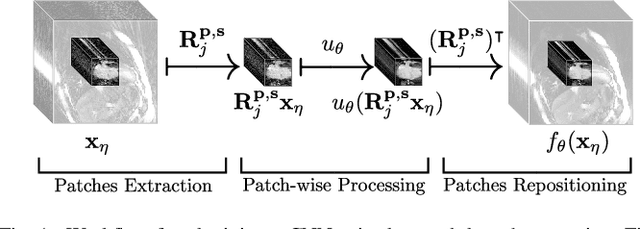
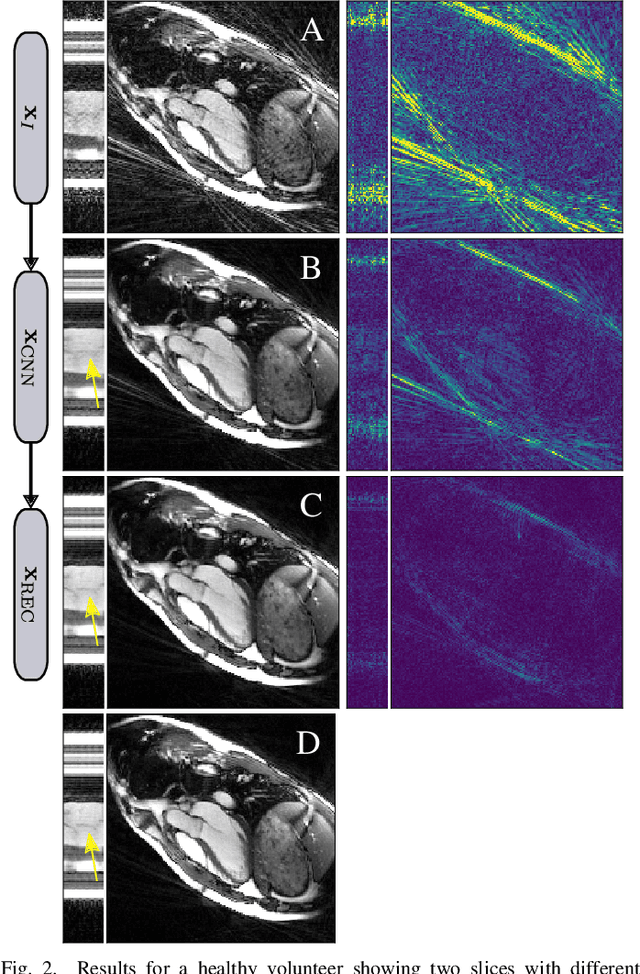

Abstract:In this paper we present a generalized Deep Learning-based approach for solving ill-posed large-scale inverse problems occuring in medical image reconstruction. Recently, Deep Learning methods using iterative neural networks and cascaded neural networks have been reported to achieve state-of-the-art results with respect to various quantitative quality measures as PSNR, NRMSE and SSIM across different imaging modalities. However, the fact that these approaches employ the forward and adjoint operators repeatedly in the network architecture requires the network to process the whole images or volumes at once, which for some applications is computationally infeasible. In this work, we follow a different reconstruction strategy by decoupling the regularization of the solution from ensuring consistency with the measured data. The regularization is given in the form of an image prior obtained by the output of a previously trained neural network which is used in a Tikhonov regularization framework. By doing so, more complex and sophisticated network architectures can be used for the removal of the artefacts or noise than it is usually the case in iterative networks. Due to the large scale of the considered problems and the resulting computational complexity of the employed networks, the priors are obtained by processing the images or volumes as patches or slices. We evaluated the method for the cases of 3D cone-beam low dose CT and undersampled 2D radial cine MRI and compared it to a total variation-minimization-based reconstruction algorithm as well as to a method with regularization based on learned overcomplete dictionaries. The proposed method outperformed all the reported methods with respect to all chosen quantitative measures and further accelerates the regularization step in the reconstruction by several orders of magnitude.
Towards Automatic Abdominal Multi-Organ Segmentation in Dual Energy CT using Cascaded 3D Fully Convolutional Network
Oct 15, 2017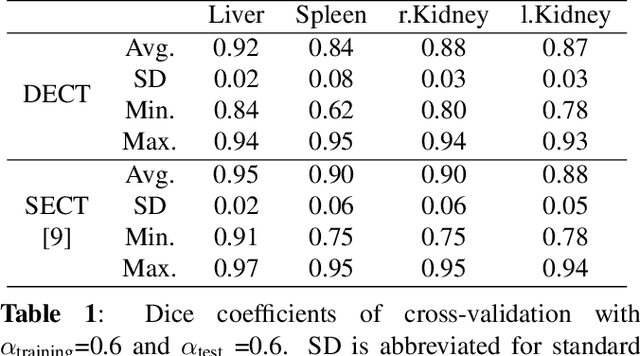
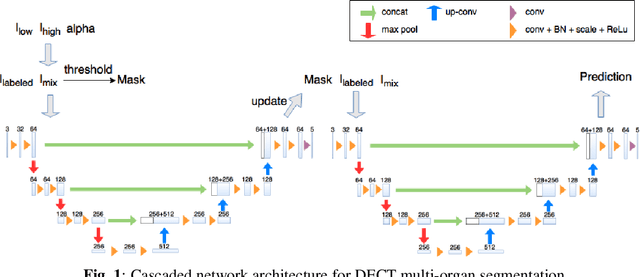
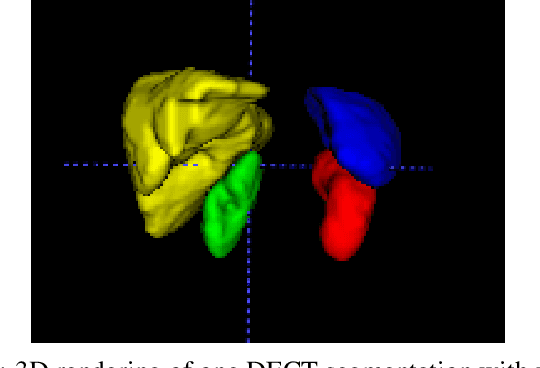
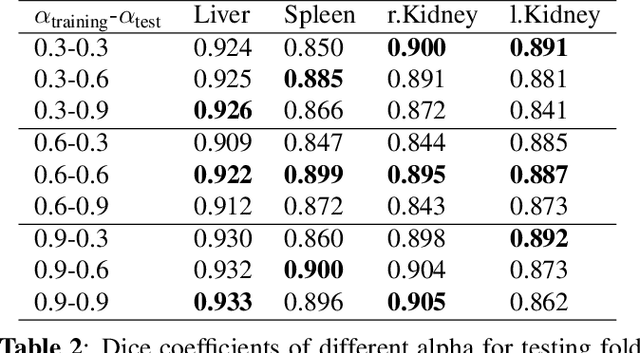
Abstract:Automatic multi-organ segmentation of the dual energy computed tomography (DECT) data can be beneficial for biomedical research and clinical applications. However, it is a challenging task. Recent advances in deep learning showed the feasibility to use 3-D fully convolutional networks (FCN) for voxel-wise dense predictions in single energy computed tomography (SECT). In this paper, we proposed a 3D FCN based method for automatic multi-organ segmentation in DECT. The work was based on a cascaded FCN and a general model for the major organs trained on a large set of SECT data. We preprocessed the DECT data by using linear weighting and fine-tuned the model for the DECT data. The method was evaluated using 42 torso DECT data acquired with a clinical dual-source CT system. Four abdominal organs (liver, spleen, left and right kidneys) were evaluated. Cross-validation was tested. Effect of the weight on the accuracy was researched. In all the tests, we achieved an average Dice coefficient of 93% for the liver, 90% for the spleen, 91% for the right kidney and 89% for the left kidney, respectively. The results show our method is feasible and promising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge