Markus Haltmeier
Schrödinger-Inspired Time-Evolution for 4D Deformation Forecasting
Jan 31, 2026Abstract:Spatiotemporal forecasting of complex three-dimensional phenomena (4D: 3D + time) is fundamental to applications in medical imaging, fluid and material dynamics, and geophysics. In contrast to unconstrained neural forecasting models, we propose a Schrödinger-inspired, physics-guided neural architecture that embeds an explicit time-evolution operator within a deep convolutional framework for 4D prediction. From observed volumetric sequences, the model learns voxelwise amplitude, phase, and potential fields that define a complex-valued wavefunction $ψ= A e^{iφ}$, which is evolved forward in time using a differentiable, unrolled Schrödinger time stepper. This physics-guided formulation yields several key advantages: (i) temporal stability arising from the structured evolution operator, which mitigates drift and error accumulation in long-horizon forecasting; (ii) an interpretable latent representation, where phase encodes transport dynamics, amplitude captures structural intensity, and the learned potential governs spatiotemporal interactions; and (iii) natural compatibility with deformation-based synthesis, which is critical for preserving anatomical fidelity in medical imaging applications. By integrating physical priors directly into the learning process, the proposed approach combines the expressivity of deep networks with the robustness and interpretability of physics-based modeling. We demonstrate accurate and stable prediction of future 4D states, including volumetric intensities and deformation fields, on synthetic benchmarks that emulate realistic shape deformations and topological changes. To our knowledge, this is the first end-to-end 4D neural forecasting framework to incorporate a Schrödinger-type evolution operator, offering a principled pathway toward interpretable, stable, and anatomically consistent spatiotemporal prediction.
Data-Consistent Learning of Inverse Problems
Jan 19, 2026Abstract:Inverse problems are inherently ill-posed, suffering from non-uniqueness and instability. Classical regularization methods provide mathematically well-founded solutions, ensuring stability and convergence, but often at the cost of reduced flexibility or visual quality. Learned reconstruction methods, such as convolutional neural networks, can produce visually compelling results, yet they typically lack rigorous theoretical guarantees. DC (DC) networks address this gap by enforcing the measurement model within the network architecture. In particular, null-space networks combined with a classical regularization method as an initial reconstruction define a convergent regularization method. This approach preserves the theoretical reliability of classical schemes while leveraging the expressive power of data-driven learning, yielding reconstructions that are both accurate and visually appealing.
HyDRA: Hybrid Denoising Regularization for Measurement-Only DEQ Training
Jan 03, 2026Abstract:Solving image reconstruction problems of the form \(\mathbf{A} \mathbf{x} = \mathbf{y}\) remains challenging due to ill-posedness and the lack of large-scale supervised datasets. Deep Equilibrium (DEQ) models have been used successfully but typically require supervised pairs \((\mathbf{x},\mathbf{y})\). In many practical settings, only measurements \(\mathbf{y}\) are available. We introduce HyDRA (Hybrid Denoising Regularization Adaptation), a measurement-only framework for DEQ training that combines measurement consistency with an adaptive denoising regularization term, together with a data-driven early stopping criterion. Experiments on sparse-view CT demonstrate competitive reconstruction quality and fast inference.
Learning Regularization Functionals for Inverse Problems: A Comparative Study
Oct 02, 2025Abstract:In recent years, a variety of learned regularization frameworks for solving inverse problems in imaging have emerged. These offer flexible modeling together with mathematical insights. The proposed methods differ in their architectural design and training strategies, making direct comparison challenging due to non-modular implementations. We address this gap by collecting and unifying the available code into a common framework. This unified view allows us to systematically compare the approaches and highlight their strengths and limitations, providing valuable insights into their future potential. We also provide concise descriptions of each method, complemented by practical guidelines.
Noisier2Inverse: Self-Supervised Learning for Image Reconstruction with Correlated Noise
Mar 25, 2025Abstract:We propose Noisier2Inverse, a correction-free self-supervised deep learning approach for general inverse prob- lems. The proposed method learns a reconstruction function without the need for ground truth samples and is ap- plicable in cases where measurement noise is statistically correlated. This includes computed tomography, where detector imperfections or photon scattering create correlated noise patterns, as well as microscopy and seismic imaging, where physical interactions during measurement introduce dependencies in the noise structure. Similar to Noisier2Noise, a key step in our approach is the generation of noisier data from which the reconstruction net- work learns. However, unlike Noisier2Noise, the proposed loss function operates in measurement space and is trained to recover an extrapolated image instead of the original noisy one. This eliminates the need for an extrap- olation step during inference, which would otherwise suffer from ill-posedness. We numerically demonstrate that our method clearly outperforms previous self-supervised approaches that account for correlated noise.
A lightweight residual network for unsupervised deformable image registration
Jun 14, 2024



Abstract:Accurate volumetric image registration is highly relevant for clinical routines and computer-aided medical diagnosis. Recently, researchers have begun to use transformers in learning-based methods for medical image registration, and have achieved remarkable success. Due to the strong global modeling capability, Transformers are considered a better option than convolutional neural networks (CNNs) for registration. However, they use bulky models with huge parameter sets, which require high computation edge devices for deployment as portable devices or in hospitals. Transformers also need a large amount of training data to produce significant results, and it is often challenging to collect suitable annotated data. Although existing CNN-based image registration can offer rich local information, their global modeling capability is poor for handling long-distance information interaction and limits registration performance. In this work, we propose a CNN-based registration method with an enhanced receptive field, a low number of parameters, and significant results on a limited training dataset. For this, we propose a residual U-Net with embedded parallel dilated-convolutional blocks to enhance the receptive field. The proposed method is evaluated on inter-patient and atlas-based datasets. We show that the performance of the proposed method is comparable and slightly better than transformer-based methods by using only $\SI{1.5}{\percent}$ of its number of parameters.
Deep Gaussian mixture model for unsupervised image segmentation
Apr 18, 2024



Abstract:The recent emergence of deep learning has led to a great deal of work on designing supervised deep semantic segmentation algorithms. As in many tasks sufficient pixel-level labels are very difficult to obtain, we propose a method which combines a Gaussian mixture model (GMM) with unsupervised deep learning techniques. In the standard GMM the pixel values with each sub-region are modelled by a Gaussian distribution. In order to identify the different regions, the parameter vector that minimizes the negative log-likelihood (NLL) function regarding the GMM has to be approximated. For this task, usually iterative optimization methods such as the expectation-maximization (EM) algorithm are used. In this paper, we propose to estimate these parameters directly from the image using a convolutional neural network (CNN). We thus change the iterative procedure in the EM algorithm replacing the expectation-step by a gradient-step with regard to the networks parameters. This means that the network is trained to minimize the NLL function of the GMM which comes with at least two advantages. As once trained, the network is able to predict label probabilities very quickly compared with time consuming iterative optimization methods. Secondly, due to the deep image prior our method is able to partially overcome one of the main disadvantages of GMM, which is not taking into account correlation between neighboring pixels, as it assumes independence between them. We demonstrate the advantages of our method in various experiments on the example of myocardial infarct segmentation on multi-sequence MRI images.
Sparse2Inverse: Self-supervised inversion of sparse-view CT data
Feb 26, 2024Abstract:Sparse-view computed tomography (CT) enables fast and low-dose CT imaging, an essential feature for patient-save medical imaging and rapid non-destructive testing. In sparse-view CT, only a few projection views are acquired, causing standard reconstructions to suffer from severe artifacts and noise. To address these issues, we propose a self-supervised image reconstruction strategy. Specifically, in contrast to the established Noise2Inverse, our proposed training strategy uses a loss function in the projection domain, thereby bypassing the otherwise prescribed nullspace component. We demonstrate the effectiveness of the proposed method in reducing stripe-artifacts and noise, even from highly sparse data.
Design, Implementation and Analysis of a Compressed Sensing Photoacoustic Projection Imaging System
Feb 24, 2024Abstract:Significance: Compressed sensing (CS) uses special measurement designs combined with powerful mathematical algorithms to reduce the amount of data to be collected while maintaining image quality. This is relevant to almost any imaging modality, and in this paper we focus on CS in photoacoustic projection imaging (PAPI) with integrating line detectors (ILDs). Aim: Our previous research involved rather general CS measurements, where each ILD can contribute to any measurement. In the real world, however, the design of CS measurements is subject to practical constraints. In this research, we aim at a CS-PAPI system where each measurement involves only a subset of ILDs, and which can be implemented in a cost-effective manner. Approach: We extend the existing PAPI with a self-developed CS unit. The system provides structured CS matrices for which the existing recovery theory cannot be applied directly. A random search strategy is applied to select the CS measurement matrix within this class for which we obtain exact sparse recovery. Results: We implement a CS PAPI system for a compression factor of $4:3$, where specific measurements are made on separate groups of 16 ILDs. We algorithmically design optimal CS measurements that have proven sparse CS capabilities. Numerical experiments are used to support our results. Conclusions: CS with proven sparse recovery capabilities can be integrated into PAPI, and numerical results support this setup. Future work will focus on applying it to experimental data and utilizing data-driven approaches to enhance the compression factor and generalize the signal class.
Self Supervised Learning for Improved Calibrationless Radial MRI with NLINV-Net
Feb 09, 2024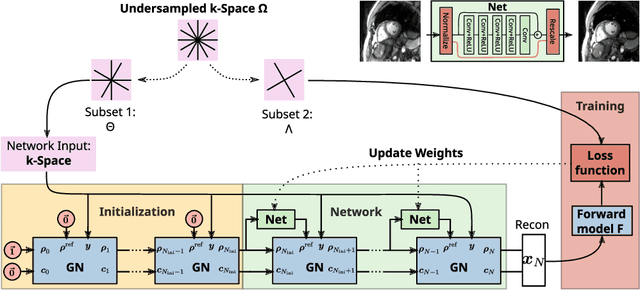
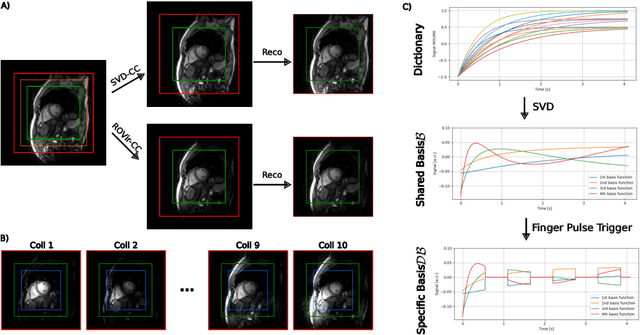
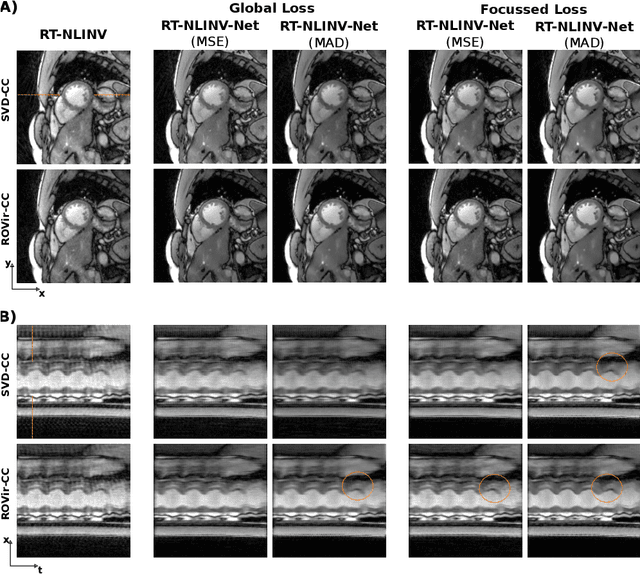
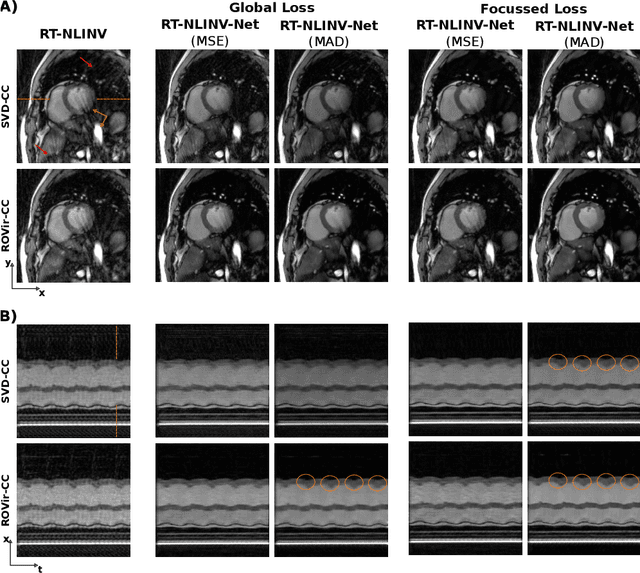
Abstract:Purpose: To develop a neural network architecture for improved calibrationless reconstruction of radial data when no ground truth is available for training. Methods: NLINV-Net is a model-based neural network architecture that directly estimates images and coil sensitivities from (radial) k-space data via non-linear inversion (NLINV). Combined with a training strategy using self-supervision via data undersampling (SSDU), it can be used for imaging problems where no ground truth reconstructions are available. We validated the method for (1) real-time cardiac imaging and (2) single-shot subspace-based quantitative T1 mapping. Furthermore, region-optimized virtual (ROVir) coils were used to suppress artifacts stemming from outside the FoV and to focus the k-space based SSDU loss on the region of interest. NLINV-Net based reconstructions were compared with conventional NLINV and PI-CS (parallel imaging + compressed sensing) reconstruction and the effect of the region-optimized virtual coils and the type of training loss was evaluated qualitatively. Results: NLINV-Net based reconstructions contain significantly less noise than the NLINV-based counterpart. ROVir coils effectively suppress streakings which are not suppressed by the neural networks while the ROVir-based focussed loss leads to visually sharper time series for the movement of the myocardial wall in cardiac real-time imaging. For quantitative imaging, T1-maps reconstructed using NLINV-Net show similar quality as PI-CS reconstructions, but NLINV-Net does not require slice-specific tuning of the regularization parameter. Conclusion: NLINV-Net is a versatile tool for calibrationless imaging which can be used in challenging imaging scenarios where a ground truth is not available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge