Christian Kremser
Deep Learning Pipeline for Fully Automated Myocardial Infarct Segmentation from Clinical Cardiac MR Scans
Feb 05, 2025



Abstract:Purpose: To develop and evaluate a deep learning-based method that allows to perform myocardial infarct segmentation in a fully-automated way. Materials and Methods: For this retrospective study, a cascaded framework of two and three-dimensional convolutional neural networks (CNNs), specialized on identifying ischemic myocardial scars on late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) images, was trained on an in-house training dataset consisting of 144 examinations. On a separate test dataset from the same institution, including images from 152 examinations obtained between 2021 and 2023, a quantitative comparison between artificial intelligence (AI)-based segmentations and manual segmentations was performed. Further, qualitative assessment of segmentation accuracy was evaluated for both human and AI-generated contours by two CMR experts in a blinded experiment. Results: Excellent agreement could be found between manually and automatically calculated infarct volumes ($\rho_c$ = 0.9). The qualitative evaluation showed that compared to human-based measurements, the experts rated the AI-based segmentations to better represent the actual extent of infarction significantly (p < 0.001) more often (33.4% AI, 25.1% human, 41.5% equal). On the contrary, for segmentation of microvascular obstruction (MVO), manual measurements were still preferred (11.3% AI, 55.6% human, 33.1% equal). Conclusion: This fully-automated segmentation pipeline enables CMR infarct size to be calculated in a very short time and without requiring any pre-processing of the input images while matching the segmentation quality of trained human observers. In a blinded experiment, experts preferred automated infarct segmentations more often than manual segmentations, paving the way for a potential clinical application.
Error correcting 2D-3D cascaded network for myocardial infarct scar segmentation on late gadolinium enhancement cardiac magnetic resonance images
Jun 26, 2023Abstract:Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) imaging is considered the in vivo reference standard for assessing infarct size (IS) and microvascular obstruction (MVO) in ST-elevation myocardial infarction (STEMI) patients. However, the exact quantification of those markers of myocardial infarct severity remains challenging and very time-consuming. As LGE distribution patterns can be quite complex and hard to delineate from the blood pool or epicardial fat, automatic segmentation of LGE CMR images is challenging. In this work, we propose a cascaded framework of two-dimensional and three-dimensional convolutional neural networks (CNNs) which enables to calculate the extent of myocardial infarction in a fully automated way. By artificially generating segmentation errors which are characteristic for 2D CNNs during training of the cascaded framework we are enforcing the detection and correction of 2D segmentation errors and hence improve the segmentation accuracy of the entire method. The proposed method was trained and evaluated in a five-fold cross validation using the training dataset from the EMIDEC challenge. We perform comparative experiments where our framework outperforms state-of-the-art methods of the EMIDEC challenge, as well as 2D and 3D nnU-Net. Furthermore, in extensive ablation studies we show the advantages that come with the proposed error correcting cascaded method.
A Joint Deep Learning Approach for Automated Liver and Tumor Segmentation
Feb 21, 2019
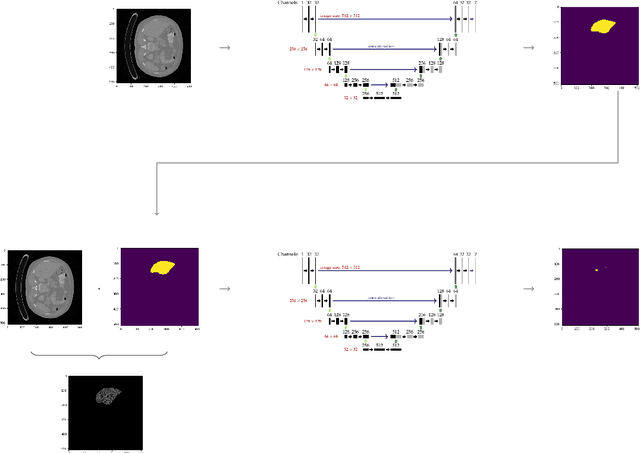
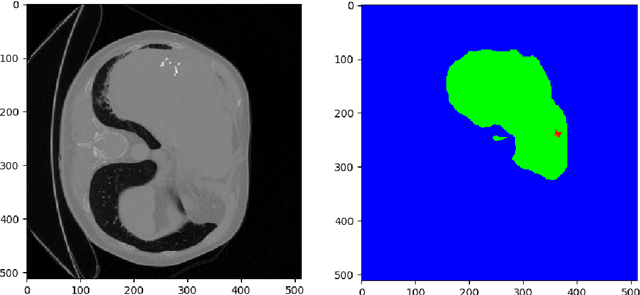
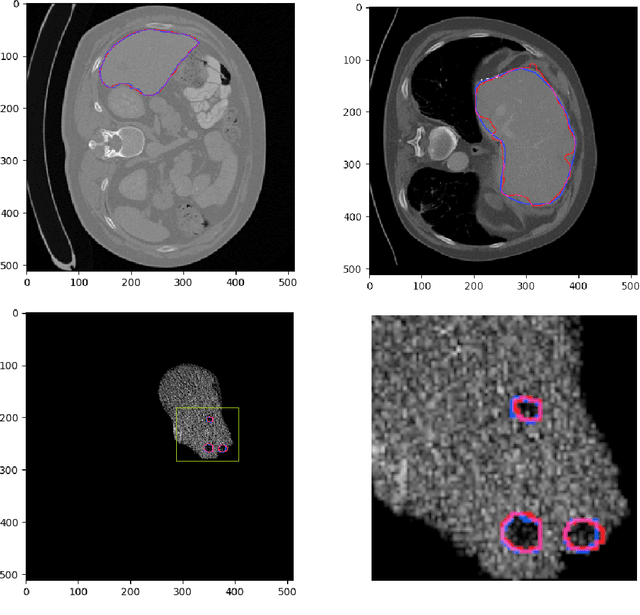
Abstract:Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer in adults, and the most common cause of death of people suffering from cirrhosis. The segmentation of liver lesions in CT images allows assessment of tumor load, treatment planning, prognosis and monitoring of treatment response. Manual segmentation is a very time-consuming task and in many cases, prone to inaccuracies and automatic tools for tumor detection and segmentation are desirable. In this paper, we use a network architecture that consists of two consecutive fully convolutional neural networks. The first network segments the liver whereas the second network segments the actual tumor inside the liver. Our network is trained on a subset of the LiTS (Liver Tumor Segmentation) challenge and evaluated on data provided from the radiological center in Innsbruck.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge