Nadja Gruber
HyDRA: Hybrid Denoising Regularization for Measurement-Only DEQ Training
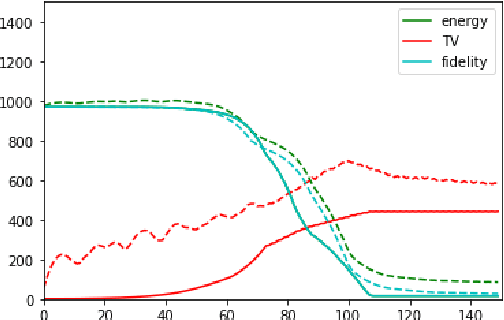
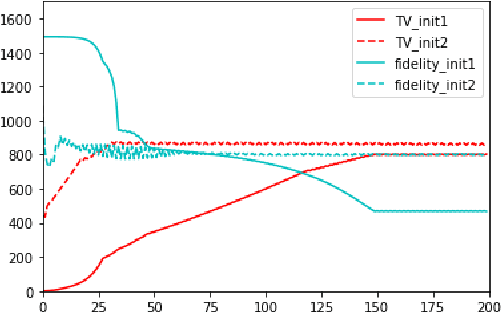
Jan 03, 2026Abstract:Solving image reconstruction problems of the form \(\mathbf{A} \mathbf{x} = \mathbf{y}\) remains challenging due to ill-posedness and the lack of large-scale supervised datasets. Deep Equilibrium (DEQ) models have been used successfully but typically require supervised pairs \((\mathbf{x},\mathbf{y})\). In many practical settings, only measurements \(\mathbf{y}\) are available. We introduce HyDRA (Hybrid Denoising Regularization Adaptation), a measurement-only framework for DEQ training that combines measurement consistency with an adaptive denoising regularization term, together with a data-driven early stopping criterion. Experiments on sparse-view CT demonstrate competitive reconstruction quality and fast inference.
Noisier2Inverse: Self-Supervised Learning for Image Reconstruction with Correlated Noise
Mar 25, 2025Abstract:We propose Noisier2Inverse, a correction-free self-supervised deep learning approach for general inverse prob- lems. The proposed method learns a reconstruction function without the need for ground truth samples and is ap- plicable in cases where measurement noise is statistically correlated. This includes computed tomography, where detector imperfections or photon scattering create correlated noise patterns, as well as microscopy and seismic imaging, where physical interactions during measurement introduce dependencies in the noise structure. Similar to Noisier2Noise, a key step in our approach is the generation of noisier data from which the reconstruction net- work learns. However, unlike Noisier2Noise, the proposed loss function operates in measurement space and is trained to recover an extrapolated image instead of the original noisy one. This eliminates the need for an extrap- olation step during inference, which would otherwise suffer from ill-posedness. We numerically demonstrate that our method clearly outperforms previous self-supervised approaches that account for correlated noise.
Sparse2Inverse: Self-supervised inversion of sparse-view CT data
Feb 26, 2024Abstract:Sparse-view computed tomography (CT) enables fast and low-dose CT imaging, an essential feature for patient-save medical imaging and rapid non-destructive testing. In sparse-view CT, only a few projection views are acquired, causing standard reconstructions to suffer from severe artifacts and noise. To address these issues, we propose a self-supervised image reconstruction strategy. Specifically, in contrast to the established Noise2Inverse, our proposed training strategy uses a loss function in the projection domain, thereby bypassing the otherwise prescribed nullspace component. We demonstrate the effectiveness of the proposed method in reducing stripe-artifacts and noise, even from highly sparse data.
Single-Image based unsupervised joint segmentation and denoising
Sep 19, 2023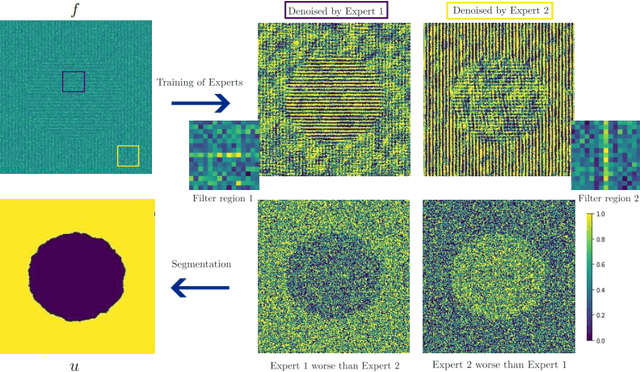

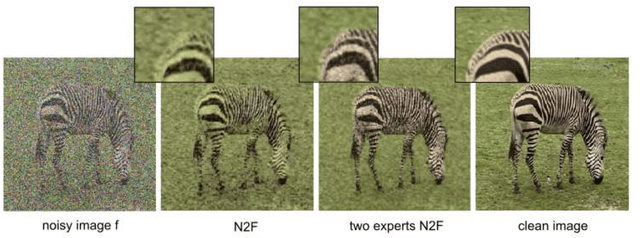
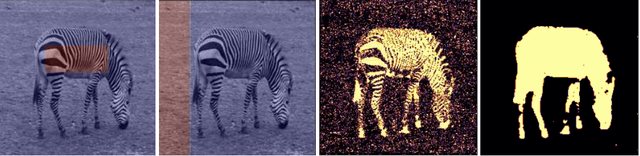
Abstract:In this work, we develop an unsupervised method for the joint segmentation and denoising of a single image. To this end, we combine the advantages of a variational segmentation method with the power of a self-supervised, single-image based deep learning approach. One major strength of our method lies in the fact, that in contrast to data-driven methods, where huge amounts of labeled samples are necessary, our model can segment an image into multiple meaningful regions without any training database. Further, we introduce a novel energy functional in which denoising and segmentation are coupled in a way that both tasks benefit from each other. The limitations of existing single-image based variational segmentation methods, which are not capable of dealing with high noise or generic texture, are tackled by this specific combination with self-supervised image denoising. We propose a unified optimisation strategy and show that, especially for very noisy images available in microscopy, our proposed joint approach outperforms its sequential counterpart as well as alternative methods focused purely on denoising or segmentation. Another comparison is conducted with a supervised deep learning approach designed for the same application, highlighting the good performance of our approach.
Variational multichannel multiclass segmentation\endgraf using unsupervised lifting with CNNs
Feb 04, 2023Abstract:We propose an unsupervised image segmentation approach, that combines a variational energy functional and deep convolutional neural networks. The variational part is based on a recent multichannel multiphase Chan-Vese model, which is capable to extract useful information from multiple input images simultaneously. We implement a flexible multiclass segmentation method that divides a given image into $K$ different regions. We use convolutional neural networks (CNNs) targeting a pre-decomposition of the image. By subsequently minimising the segmentation functional, the final segmentation is obtained in a fully unsupervised manner. Special emphasis is given to the extraction of informative feature maps serving as a starting point for the segmentation. The initial results indicate that the proposed method is able to decompose and segment the different regions of various types of images, such as texture and medical images and compare its performance with another multiphase segmentation method.
A Joint Variational Multichannel Multiphase Segmentation Framework
Feb 09, 2022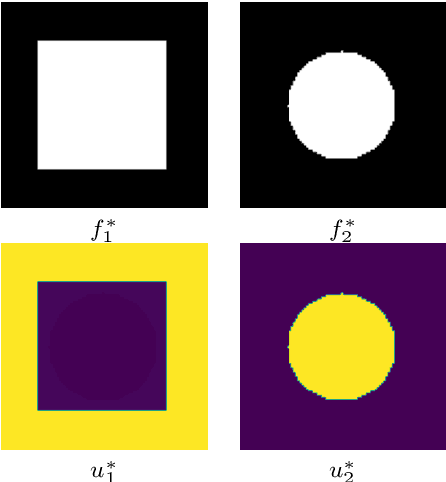
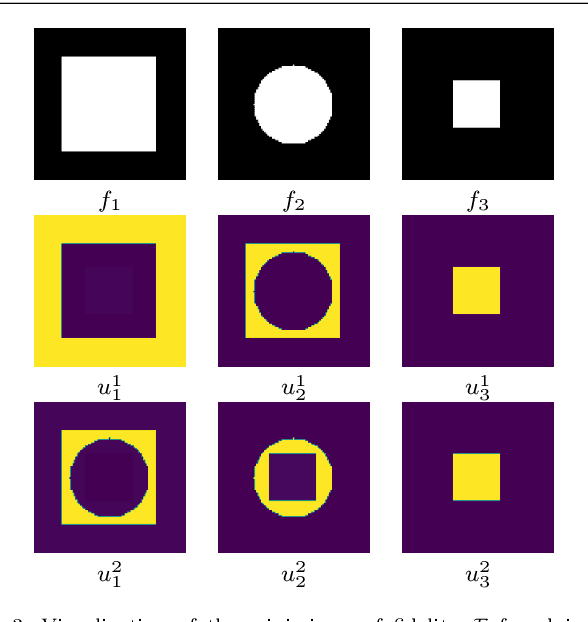
Abstract:In this paper, we propose a variational image segmentation framework for multichannel multiphase image segmentation based on the Chan-Vese active contour model. The core of our method lies in finding a variable u encoding the segmentation, by minimizing a multichannel energy functional that combines the information of multiple images. We create a decomposition of the input, either by multichannel filtering, or simply by using plain natural RGB, or medical images, which already consist of several channels. Subsequently we minimize the proposed functional for each of the channels simultaneously. Our model meets the necessary assumptions such that it can be solved efficiently by optimization techniques like the Chambolle-Pock method. We prove that the proposed energy functional has global minimizers, and show its stability and convergence with respect to noisy inputs. Experimental results show that the proposed method performs well in single- and multichannel segmentation tasks, and can be employed to the segmentation of various types of images, such as natural and texture images as well as medical images.
A Joint Deep Learning Approach for Automated Liver and Tumor Segmentation
Feb 21, 2019
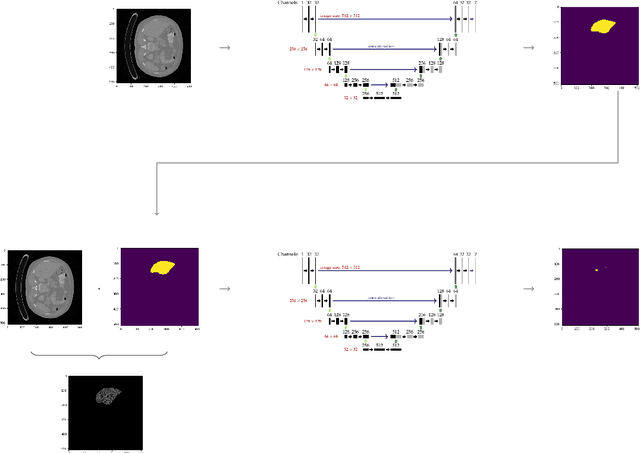
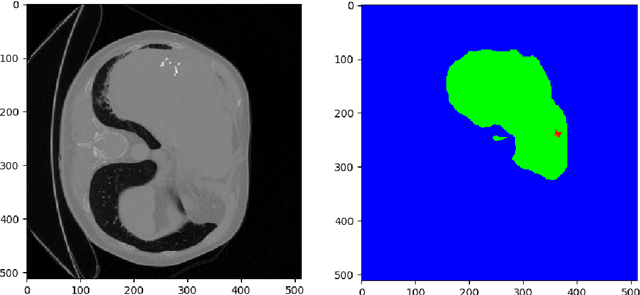
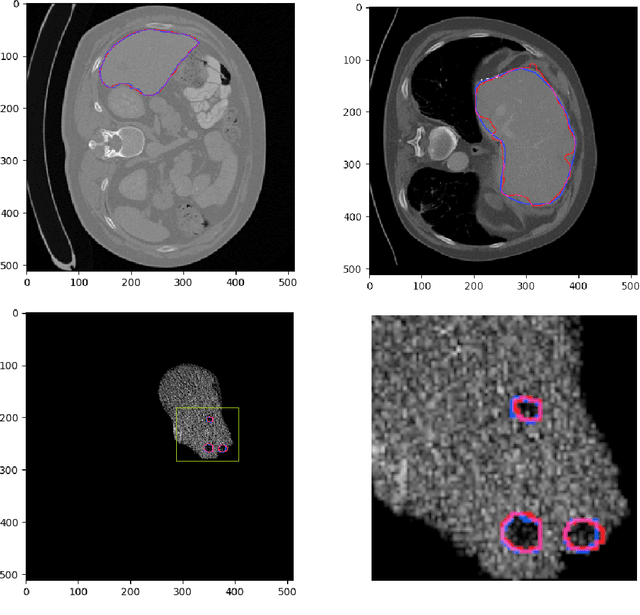
Abstract:Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer in adults, and the most common cause of death of people suffering from cirrhosis. The segmentation of liver lesions in CT images allows assessment of tumor load, treatment planning, prognosis and monitoring of treatment response. Manual segmentation is a very time-consuming task and in many cases, prone to inaccuracies and automatic tools for tumor detection and segmentation are desirable. In this paper, we use a network architecture that consists of two consecutive fully convolutional neural networks. The first network segments the liver whereas the second network segments the actual tumor inside the liver. Our network is trained on a subset of the LiTS (Liver Tumor Segmentation) challenge and evaluated on data provided from the radiological center in Innsbruck.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge