Shuqing Chen
Action Learning for 3D Point Cloud Based Organ Segmentation
Jun 14, 2018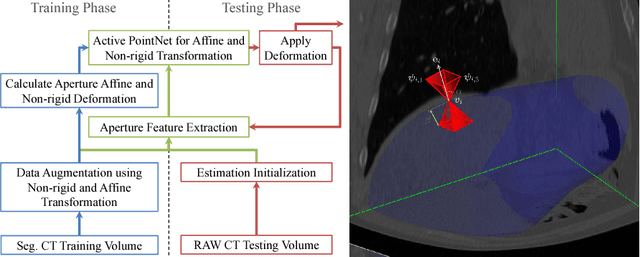
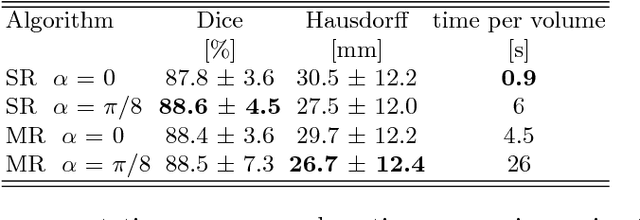
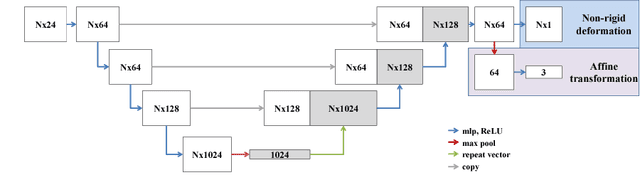
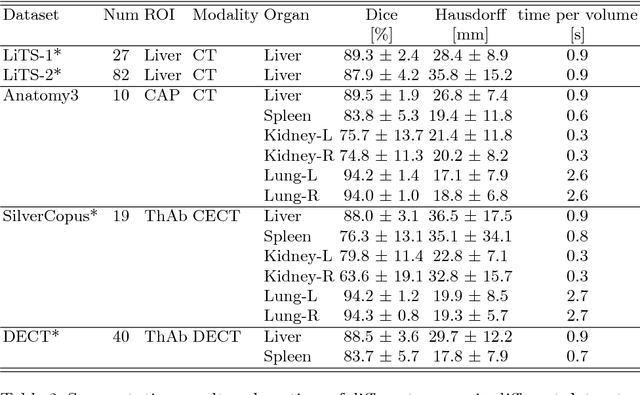
Abstract:We propose a novel point cloud based 3D organ segmentation pipeline utilizing deep Q-learning. In order to preserve shape properties, the learning process is guided using a statistical shape model. The trained agent directly predicts piece-wise linear transformations for all vertices in each iteration. This mapping between the ideal transformation for an object outline estimation is learned based on image features. To this end, we introduce aperture features that extract gray values by sampling the 3D volume within the cone centered around the associated vertex and its normal vector. Our approach is also capable of estimating a hierarchical pyramid of non rigid deformations for multi-resolution meshes. In the application phase, we use a marginal approach to gradually estimate affine as well as non-rigid transformations. We performed extensive evaluations to highlight the robust performance of our approach on a variety of challenge data as well as clinical data. Additionally, our method has a run time ranging from 0.3 to 2.7 seconds to segment each organ. In addition, we show that the proposed method can be applied to different organs, X-ray based modalities, and scanning protocols without the need of transfer learning. As we learn actions, even unseen reference meshes can be processed as demonstrated in an example with the Visible Human. From this we conclude that our method is robust, and we believe that our method can be successfully applied to many more applications, in particular, in the interventional imaging space.
Towards Automatic Abdominal Multi-Organ Segmentation in Dual Energy CT using Cascaded 3D Fully Convolutional Network
Oct 15, 2017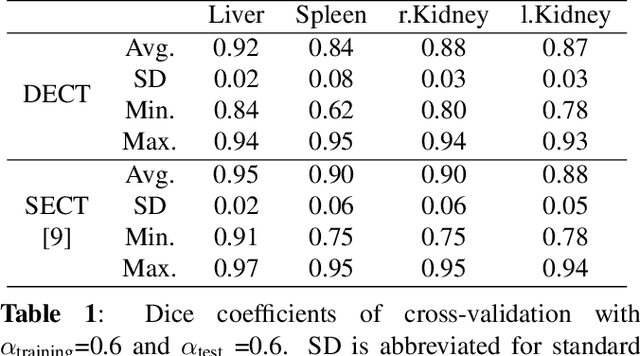
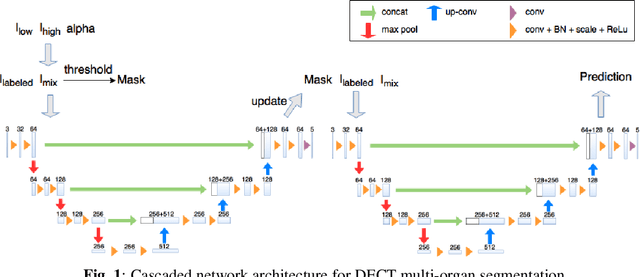
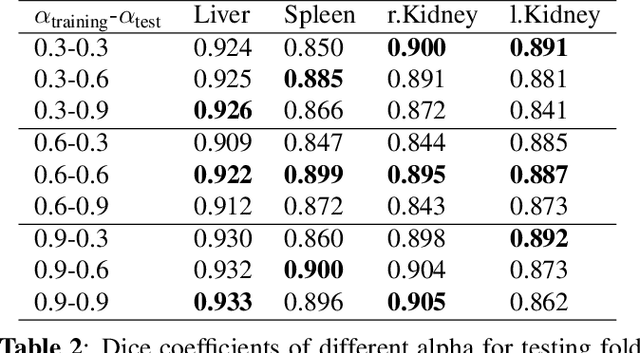
Abstract:Automatic multi-organ segmentation of the dual energy computed tomography (DECT) data can be beneficial for biomedical research and clinical applications. However, it is a challenging task. Recent advances in deep learning showed the feasibility to use 3-D fully convolutional networks (FCN) for voxel-wise dense predictions in single energy computed tomography (SECT). In this paper, we proposed a 3D FCN based method for automatic multi-organ segmentation in DECT. The work was based on a cascaded FCN and a general model for the major organs trained on a large set of SECT data. We preprocessed the DECT data by using linear weighting and fine-tuned the model for the DECT data. The method was evaluated using 42 torso DECT data acquired with a clinical dual-source CT system. Four abdominal organs (liver, spleen, left and right kidneys) were evaluated. Cross-validation was tested. Effect of the weight on the accuracy was researched. In all the tests, we achieved an average Dice coefficient of 93% for the liver, 90% for the spleen, 91% for the right kidney and 89% for the left kidney, respectively. The results show our method is feasible and promising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge