Markus Kowarschik
Siemens Healthcare GmbH, Forchheim, Germany
Advancing Embodied Intelligence in Robotic-Assisted Endovascular Procedures: A Systematic Review of AI Solutions
Apr 21, 2025Abstract:Endovascular procedures have revolutionized the treatment of vascular diseases thanks to minimally invasive solutions that significantly reduce patient recovery time and enhance clinical outcomes. However, the precision and dexterity required during these procedures poses considerable challenges for interventionists. Robotic systems have emerged offering transformative solutions, addressing issues such as operator fatigue, radiation exposure, and the inherent limitations of human precision. The integration of Embodied Intelligence (EI) into these systems signifies a paradigm shift, enabling robots to navigate complex vascular networks and adapt to dynamic physiological conditions. Data-driven approaches, advanced computer vision, medical image analysis, and machine learning techniques, are at the forefront of this evolution. These methods augment procedural intelligence by facilitating real-time vessel segmentation, device tracking, and anatomical landmark detection. Reinforcement learning and imitation learning further refine navigation strategies and replicate experts' techniques. This review systematically examines the integration of EI principles into robotic technologies, in relation to endovascular procedures. We discuss recent advancements in intelligent perception and data-driven control, and their practical applications in robot-assisted endovascular procedures. By critically evaluating current limitations and emerging opportunities, this review establishes a framework for future developments, emphasizing the potential for greater autonomy and improved clinical outcomes. Emerging trends and specific areas of research, such as federated learning for medical data sharing, explainable AI for clinical decision support, and advanced human-robot collaboration paradigms, are also explored, offering insights into the future direction of this rapidly evolving field.
ProPLIKS: Probablistic 3D human body pose estimation
Dec 05, 2024
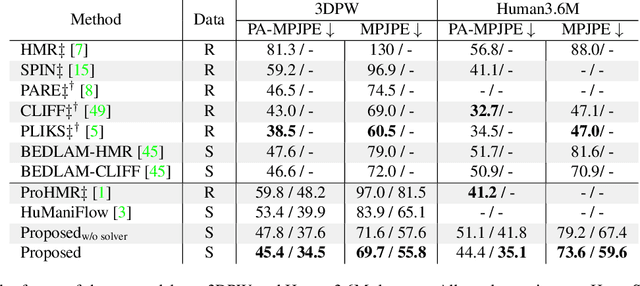

Abstract:We present a novel approach for 3D human pose estimation by employing probabilistic modeling. This approach leverages the advantages of normalizing flows in non-Euclidean geometries to address uncertain poses. Specifically, our method employs normalizing flow tailored to the SO(3) rotational group, incorporating a coupling mechanism based on the M\"obius transformation. This enables the framework to accurately represent any distribution on SO(3), effectively addressing issues related to discontinuities. Additionally, we reinterpret the challenge of reconstructing 3D human figures from 2D pixel-aligned inputs as the task of mapping these inputs to a range of probable poses. This perspective acknowledges the intrinsic ambiguity of the task and facilitates a straightforward integration method for multi-view scenarios. The combination of these strategies showcases the effectiveness of probabilistic models in complex scenarios for human pose estimation techniques. Our approach notably surpasses existing methods in the field of pose estimation. We also validate our methodology on human pose estimation from RGB images as well as medical X-Ray datasets.
Physics-Informed Learning for Time-Resolved Angiographic Contrast Agent Concentration Reconstruction
Mar 04, 2024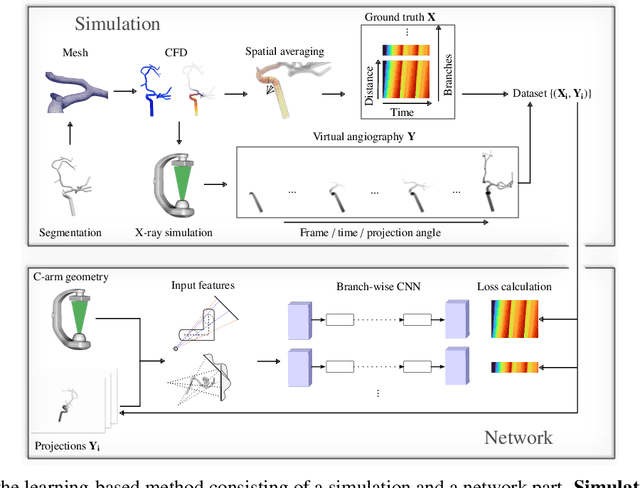
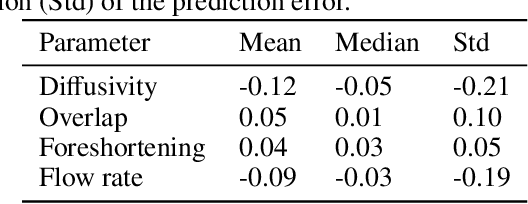
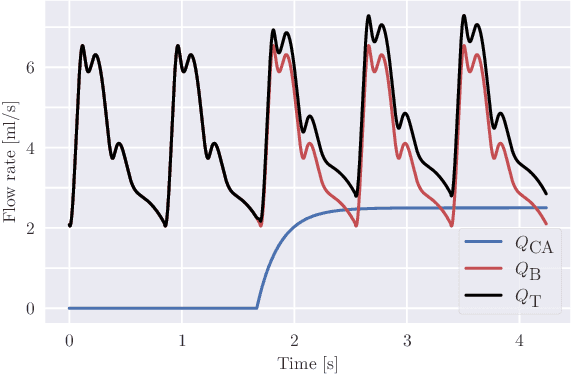
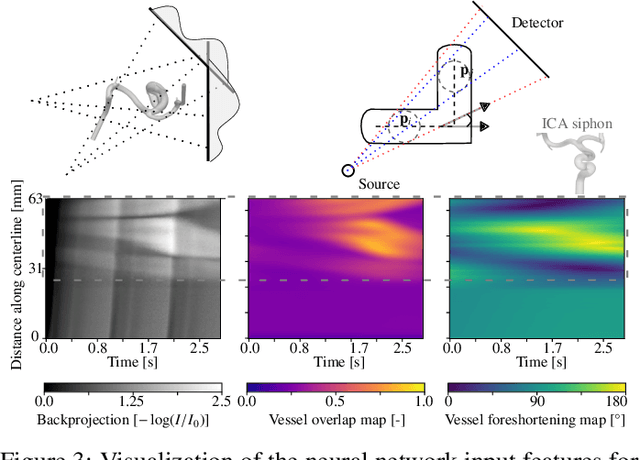
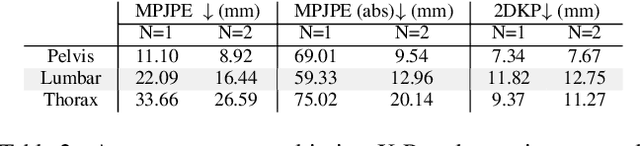
Abstract:Three-dimensional Digital Subtraction Angiography (3D-DSA) is a well-established X-ray-based technique for visualizing vascular anatomy. Recently, four-dimensional DSA (4D-DSA) reconstruction algorithms have been developed to enable the visualization of volumetric contrast flow dynamics through time-series of volumes. . This reconstruction problem is ill-posed mainly due to vessel overlap in the projection direction and geometric vessel foreshortening, which leads to information loss in the recorded projection images. However, knowledge about the underlying fluid dynamics can be leveraged to constrain the solution space. In our work, we implicitly include this information in a neural network-based model that is trained on a dataset of image-based blood flow simulations. The model predicts the spatially averaged contrast agent concentration for each centerline point of the vasculature over time, lowering the overall computational demand. The trained network enables the reconstruction of relative contrast agent concentrations with a mean absolute error of 0.02 $\pm$ 0.02 and a mean absolute percentage error of 5.31 % $\pm$ 9.25 %. Moreover, the network is robust to varying degrees of vessel overlap and vessel foreshortening. Our approach demonstrates the potential of the integration of machine learning and blood flow simulations in time-resolved angiographic flow reconstruction.
BOSS: Bones, Organs and Skin Shape Model
Mar 08, 2023Abstract:Objective: A digital twin of a patient can be a valuable tool for enhancing clinical tasks such as workflow automation, patient-specific X-ray dose optimization, markerless tracking, positioning, and navigation assistance in image-guided interventions. However, it is crucial that the patient's surface and internal organs are of high quality for any pose and shape estimates. At present, the majority of statistical shape models (SSMs) are restricted to a small number of organs or bones or do not adequately represent the general population. Method: To address this, we propose a deformable human shape and pose model that combines skin, internal organs, and bones, learned from CT images. By modeling the statistical variations in a pose-normalized space using probabilistic PCA while also preserving joint kinematics, our approach offers a holistic representation of the body that can benefit various medical applications. Results: We assessed our model's performance on a registered dataset, utilizing the unified shape space, and noted an average error of 3.6 mm for bones and 8.8 mm for organs. To further verify our findings, we conducted additional tests on publicly available datasets with multi-part segmentations, which confirmed the effectiveness of our model. Conclusion: This works shows that anatomically parameterized statistical shape models can be created accurately and in a computationally efficient manner. Significance: The proposed approach enables the construction of shape models that can be directly applied to various medical applications, including biomechanics and reconstruction.
Transient Hemodynamics Prediction Using an Efficient Octree-Based Deep Learning Model
Feb 13, 2023
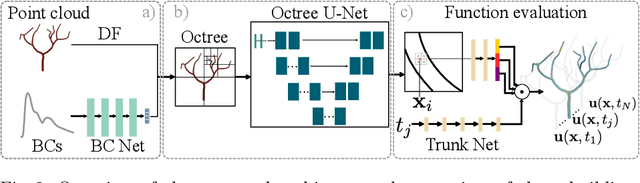
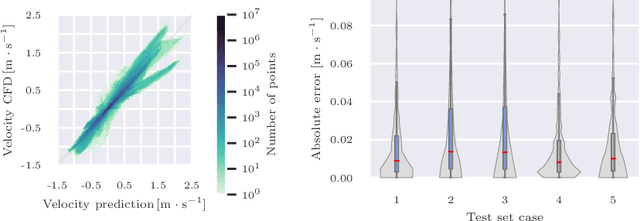
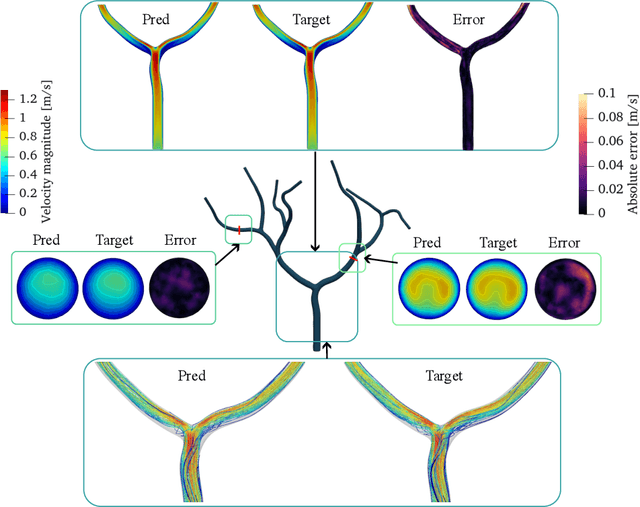
Abstract:Patient-specific hemodynamics assessment could support diagnosis and treatment of neurovascular diseases. Currently, conventional medical imaging modalities are not able to accurately acquire high-resolution hemodynamic information that would be required to assess complex neurovascular pathologies. Therefore, computational fluid dynamics (CFD) simulations can be applied to tomographic reconstructions to obtain clinically relevant information. However, three-dimensional (3D) CFD simulations require enormous computational resources and simulation-related expert knowledge that are usually not available in clinical environments. Recently, deep-learning-based methods have been proposed as CFD surrogates to improve computational efficiency. Nevertheless, the prediction of high-resolution transient CFD simulations for complex vascular geometries poses a challenge to conventional deep learning models. In this work, we present an architecture that is tailored to predict high-resolution (spatial and temporal) velocity fields for complex synthetic vascular geometries. For this, an octree-based spatial discretization is combined with an implicit neural function representation to efficiently handle the prediction of the 3D velocity field for each time step. The presented method is evaluated for the task of cerebral hemodynamics prediction before and during the injection of contrast agent in the internal carotid artery (ICA). Compared to CFD simulations, the velocity field can be estimated with a mean absolute error of 0.024 m/s, whereas the run time reduces from several hours on a high-performance cluster to a few seconds on a consumer graphical processing unit.
PLIKS: A Pseudo-Linear Inverse Kinematic Solver for 3D Human Body Estimation
Nov 21, 2022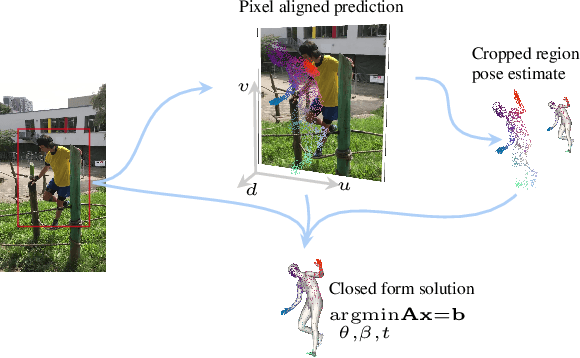
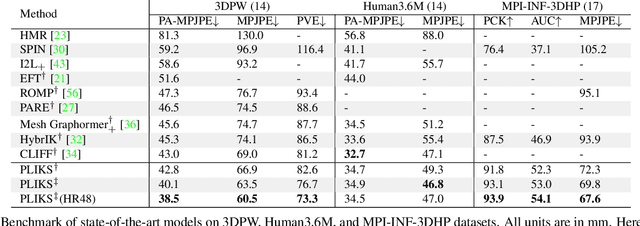
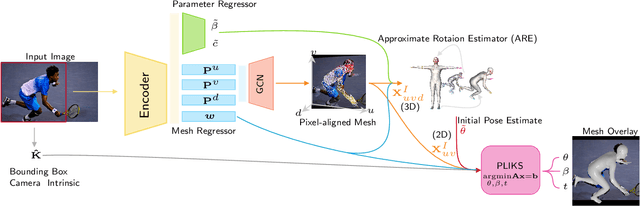
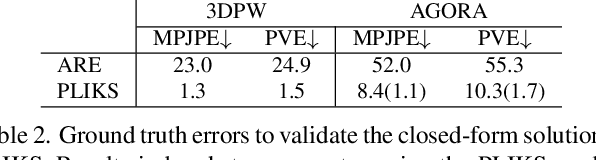
Abstract:We consider the problem of reconstructing a 3D mesh of the human body from a single 2D image as a model-in-the-loop optimization problem. Existing approaches often regress the shape, pose, and translation parameters of a parametric statistical model assuming a weak-perspective camera. In contrast, we first estimate 2D pixel-aligned vertices in image space and propose PLIKS (Pseudo-Linear Inverse Kinematic Solver) to regress the model parameters by minimizing a linear least squares problem. PLIKS is a linearized formulation of the parametric SMPL model, which provides an optimal pose and shape solution from an adequate initialization. Our method is based on analytically calculating an initial pose estimate from the network predicted 3D mesh followed by PLIKS to obtain an optimal solution for the given constraints. As our framework makes use of 2D pixel-aligned maps, it is inherently robust to partial occlusion. To demonstrate the performance of the proposed approach, we present quantitative evaluations which confirm that PLIKS achieves more accurate reconstruction with greater than 10% improvement compared to other state-of-the-art methods with respect to the standard 3D human pose and shape benchmarks while also obtaining a reconstruction error improvement of 12.9 mm on the newer AGORA dataset.
Deep Learning compatible Differentiable X-ray Projections for Inverse Rendering
Feb 04, 2021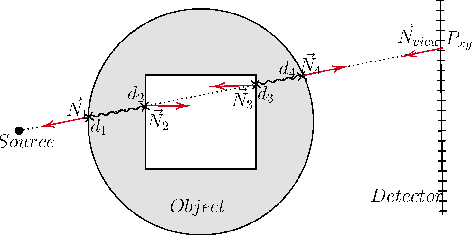
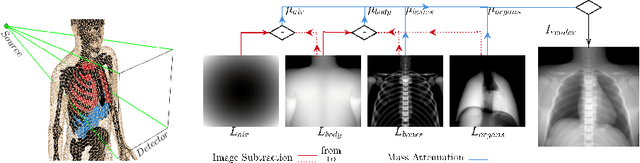

Abstract:Many minimally invasive interventional procedures still rely on 2D fluoroscopic imaging. Generating a patient-specific 3D model from these X-ray projection data would allow to improve the procedural workflow, e.g. by providing assistance functions such as automatic positioning. To accomplish this, two things are required. First, a statistical human shape model of the human anatomy and second, a differentiable X-ray renderer. In this work, we propose a differentiable renderer by deriving the distance travelled by a ray inside mesh structures to generate a distance map. To demonstrate its functioning, we use it for simulating X-ray images from human shape models. Then we show its application by solving the inverse problem, namely reconstructing 3D models from real 2D fluoroscopy images of the pelvis, which is an ideal anatomical structure for patient registration. This is accomplished by an iterative optimization strategy using gradient descent. With the majority of the pelvis being in the fluoroscopic field of view, we achieve a mean Hausdorff distance of 30 mm between the reconstructed model and the ground truth segmentation.
Simultaneous Estimation of X-ray Back-Scatter and Forward-Scatter using Multi-Task Learning
Jul 08, 2020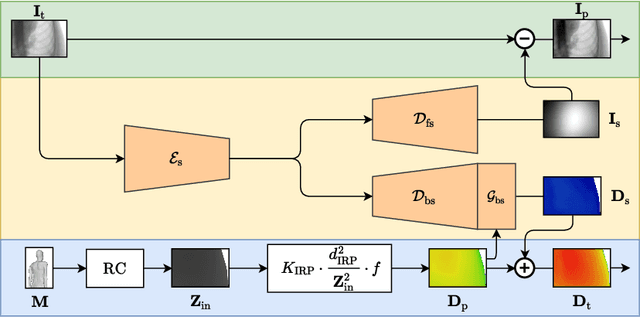
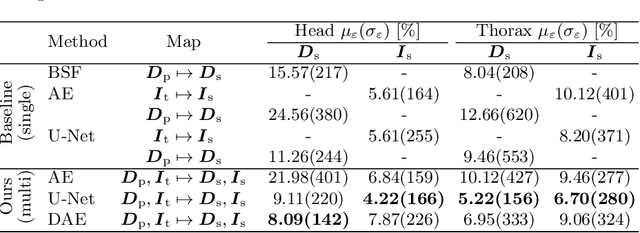
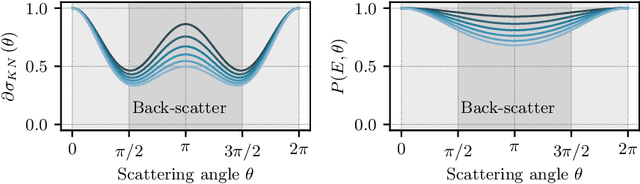
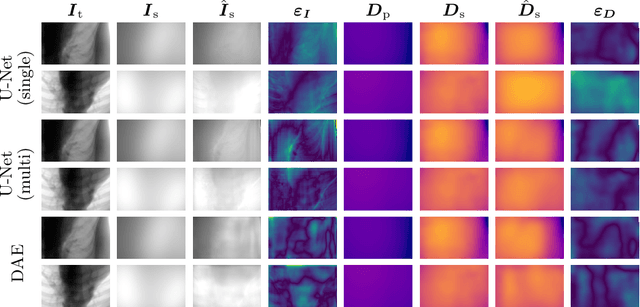
Abstract:Scattered radiation is a major concern impacting X-ray image-guided procedures in two ways. First, back-scatter significantly contributes to patient (skin) dose during complicated interventions. Second, forward-scattered radiation reduces contrast in projection images and introduces artifacts in 3-D reconstructions. While conventionally employed anti-scatter grids improve image quality by blocking X-rays, the additional attenuation due to the anti-scatter grid at the detector needs to be compensated for by a higher patient entrance dose. This also increases the room dose affecting the staff caring for the patient. For skin dose quantification, back-scatter is usually accounted for by applying pre-determined scalar back-scatter factors or linear point spread functions to a primary kerma forward projection onto a patient surface point. However, as patients come in different shapes, the generalization of conventional methods is limited. Here, we propose a novel approach combining conventional techniques with learning-based methods to simultaneously estimate the forward-scatter reaching the detector as well as the back-scatter affecting the patient skin dose. Knowing the forward-scatter, we can correct X-ray projections, while a good estimate of the back-scatter component facilitates an improved skin dose assessment. To simultaneously estimate forward-scatter as well as back-scatter, we propose a multi-task approach for joint back- and forward-scatter estimation by combining X-ray physics with neural networks. We show that, in theory, highly accurate scatter estimation in both cases is possible. In addition, we identify research directions for our multi-task framework and learning-based scatter estimation in general.
Appearance Learning for Image-based Motion Estimation in Tomography
Jun 18, 2020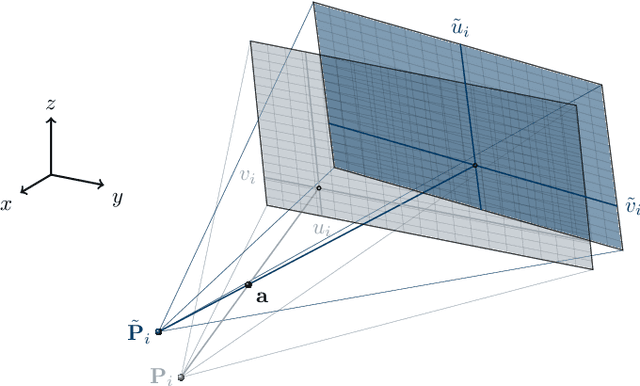
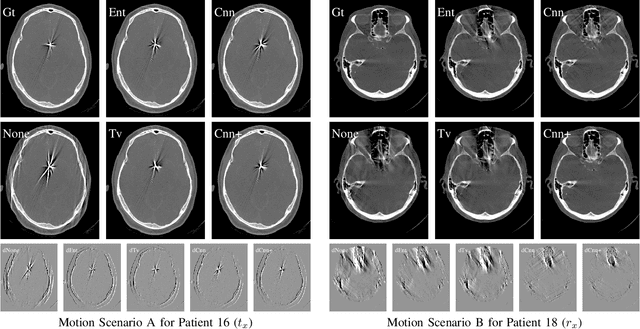
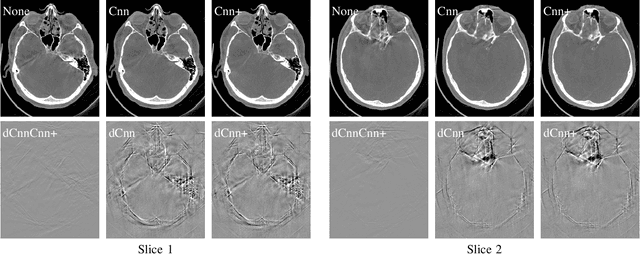
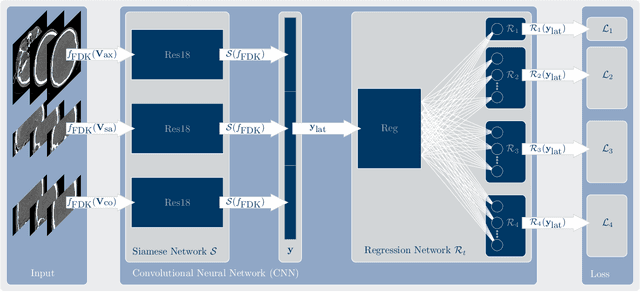
Abstract:In tomographic imaging, anatomical structures are reconstructed by applying a pseudo-inverse forward model to acquired signals. Geometric information within this process is usually depending on the system setting only, i. e., the scanner position or readout direction. Patient motion therefore corrupts the geometry alignment in the reconstruction process resulting in motion artifacts. We propose an appearance learning approach recognizing the structures of rigid motion independently from the scanned object. To this end, we train a siamese triplet network to predict the reprojection error (RPE) for the complete acquisition as well as an approximate distribution of the RPE along the single views from the reconstructed volume in a multi-task learning approach. The RPE measures the motioninduced geometric deviations independent of the object based on virtual marker positions, which are available during training. We train our network using 27 patients and deploy a 21-4-2 split for training, validation and testing. In average, we achieve a residual mean RPE of 0.013mm with an inter-patient standard deviation of 0.022 mm. This is twice the accuracy compared to previously published results. In a motion estimation benchmark the proposed approach achieves superior results in comparison with two state-of-the-art measures in nine out of twelve experiments. The clinical applicability of the proposed method is demonstrated on a motion-affected clinical dataset.
Deep autofocus with cone-beam CT consistency constraint
Dec 04, 2019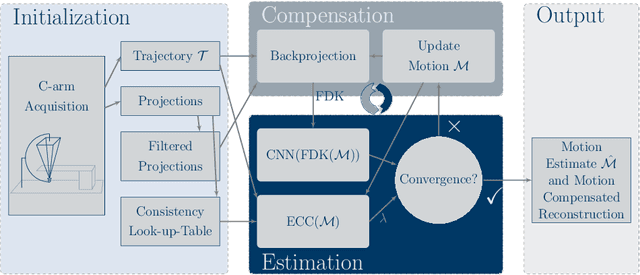
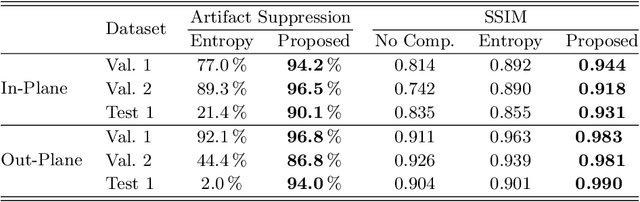
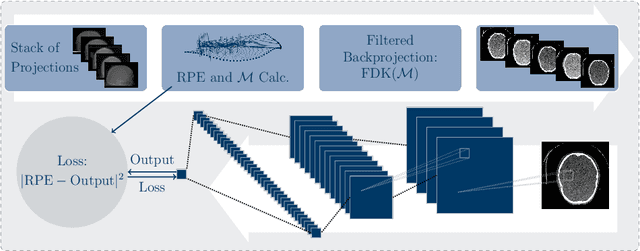
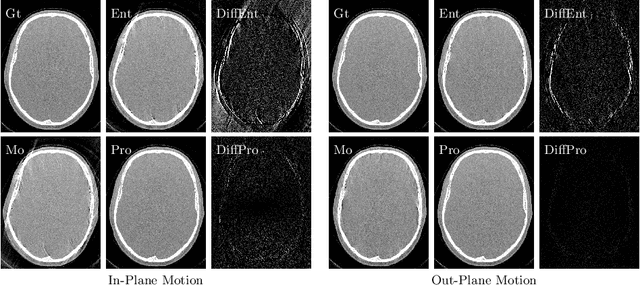
Abstract:High quality reconstruction with interventional C-arm cone-beam computed tomography (CBCT) requires exact geometry information. If the geometry information is corrupted, e. g., by unexpected patient or system movement, the measured signal is misplaced in the backprojection operation. With prolonged acquisition times of interventional C-arm CBCT the likelihood of rigid patient motion increases. To adapt the backprojection operation accordingly, a motion estimation strategy is necessary. Recently, a novel learning-based approach was proposed, capable of compensating motions within the acquisition plane. We extend this method by a CBCT consistency constraint, which was proven to be efficient for motions perpendicular to the acquisition plane. By the synergistic combination of these two measures, in and out-plane motion is well detectable, achieving an average artifact suppression of 93 [percent]. This outperforms the entropy-based state-of-the-art autofocus measure which achieves on average an artifact suppression of 54 [percent].
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge