Karthik Shetty
ProPLIKS: Probablistic 3D human body pose estimation
Dec 05, 2024
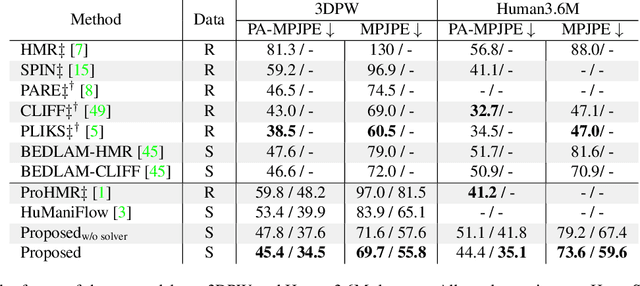

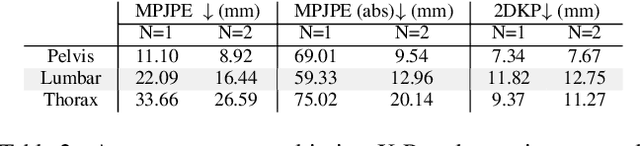
Abstract:We present a novel approach for 3D human pose estimation by employing probabilistic modeling. This approach leverages the advantages of normalizing flows in non-Euclidean geometries to address uncertain poses. Specifically, our method employs normalizing flow tailored to the SO(3) rotational group, incorporating a coupling mechanism based on the M\"obius transformation. This enables the framework to accurately represent any distribution on SO(3), effectively addressing issues related to discontinuities. Additionally, we reinterpret the challenge of reconstructing 3D human figures from 2D pixel-aligned inputs as the task of mapping these inputs to a range of probable poses. This perspective acknowledges the intrinsic ambiguity of the task and facilitates a straightforward integration method for multi-view scenarios. The combination of these strategies showcases the effectiveness of probabilistic models in complex scenarios for human pose estimation techniques. Our approach notably surpasses existing methods in the field of pose estimation. We also validate our methodology on human pose estimation from RGB images as well as medical X-Ray datasets.
BOSS: Bones, Organs and Skin Shape Model
Mar 08, 2023Abstract:Objective: A digital twin of a patient can be a valuable tool for enhancing clinical tasks such as workflow automation, patient-specific X-ray dose optimization, markerless tracking, positioning, and navigation assistance in image-guided interventions. However, it is crucial that the patient's surface and internal organs are of high quality for any pose and shape estimates. At present, the majority of statistical shape models (SSMs) are restricted to a small number of organs or bones or do not adequately represent the general population. Method: To address this, we propose a deformable human shape and pose model that combines skin, internal organs, and bones, learned from CT images. By modeling the statistical variations in a pose-normalized space using probabilistic PCA while also preserving joint kinematics, our approach offers a holistic representation of the body that can benefit various medical applications. Results: We assessed our model's performance on a registered dataset, utilizing the unified shape space, and noted an average error of 3.6 mm for bones and 8.8 mm for organs. To further verify our findings, we conducted additional tests on publicly available datasets with multi-part segmentations, which confirmed the effectiveness of our model. Conclusion: This works shows that anatomically parameterized statistical shape models can be created accurately and in a computationally efficient manner. Significance: The proposed approach enables the construction of shape models that can be directly applied to various medical applications, including biomechanics and reconstruction.
PLIKS: A Pseudo-Linear Inverse Kinematic Solver for 3D Human Body Estimation
Nov 21, 2022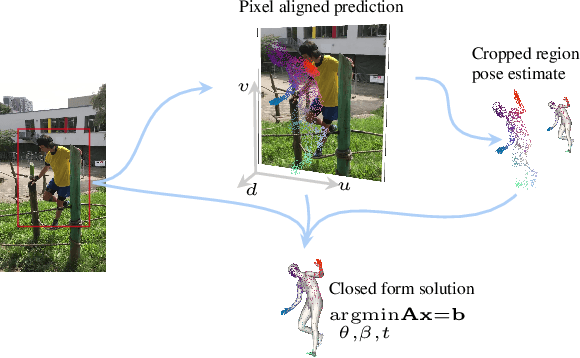
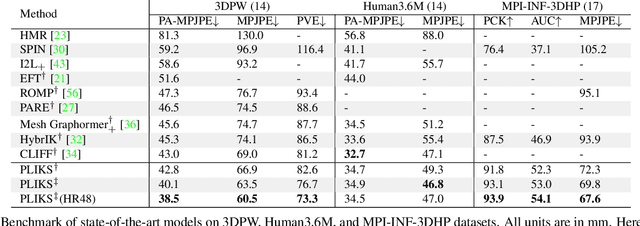
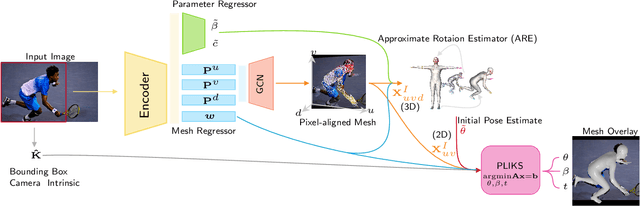
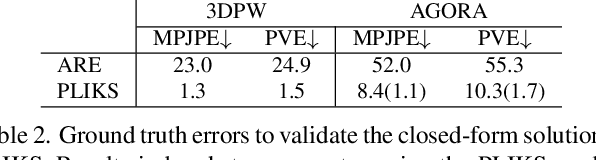
Abstract:We consider the problem of reconstructing a 3D mesh of the human body from a single 2D image as a model-in-the-loop optimization problem. Existing approaches often regress the shape, pose, and translation parameters of a parametric statistical model assuming a weak-perspective camera. In contrast, we first estimate 2D pixel-aligned vertices in image space and propose PLIKS (Pseudo-Linear Inverse Kinematic Solver) to regress the model parameters by minimizing a linear least squares problem. PLIKS is a linearized formulation of the parametric SMPL model, which provides an optimal pose and shape solution from an adequate initialization. Our method is based on analytically calculating an initial pose estimate from the network predicted 3D mesh followed by PLIKS to obtain an optimal solution for the given constraints. As our framework makes use of 2D pixel-aligned maps, it is inherently robust to partial occlusion. To demonstrate the performance of the proposed approach, we present quantitative evaluations which confirm that PLIKS achieves more accurate reconstruction with greater than 10% improvement compared to other state-of-the-art methods with respect to the standard 3D human pose and shape benchmarks while also obtaining a reconstruction error improvement of 12.9 mm on the newer AGORA dataset.
Self-Supervised 2D/3D Registration for X-Ray to CT Image Fusion
Oct 14, 2022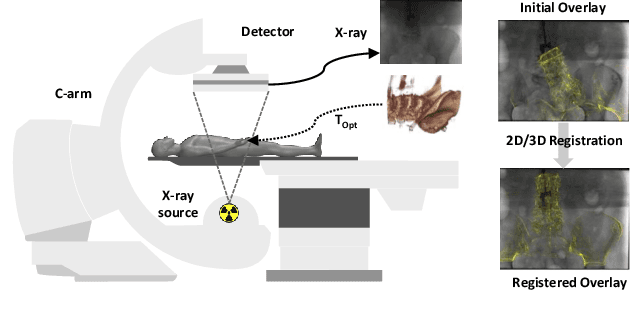

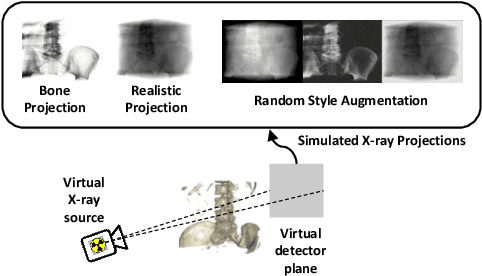
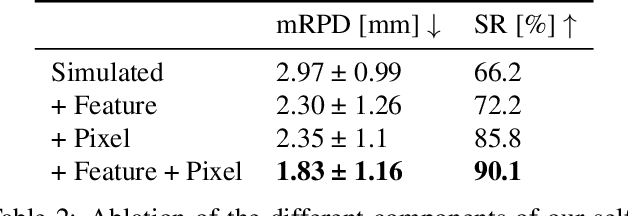
Abstract:Deep Learning-based 2D/3D registration enables fast, robust, and accurate X-ray to CT image fusion when large annotated paired datasets are available for training. However, the need for paired CT volume and X-ray images with ground truth registration limits the applicability in interventional scenarios. An alternative is to use simulated X-ray projections from CT volumes, thus removing the need for paired annotated datasets. Deep Neural Networks trained exclusively on simulated X-ray projections can perform significantly worse on real X-ray images due to the domain gap. We propose a self-supervised 2D/3D registration framework combining simulated training with unsupervised feature and pixel space domain adaptation to overcome the domain gap and eliminate the need for paired annotated datasets. Our framework achieves a registration accuracy of 1.83$\pm$1.16 mm with a high success ratio of 90.1% on real X-ray images showing a 23.9% increase in success ratio compared to reference annotation-free algorithms.
Deep Iterative 2D/3D Registration
Jul 21, 2021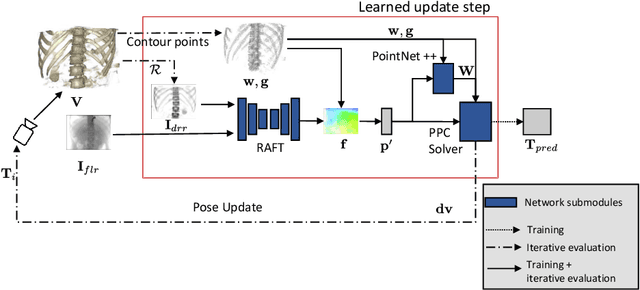
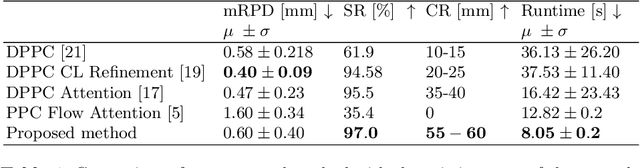
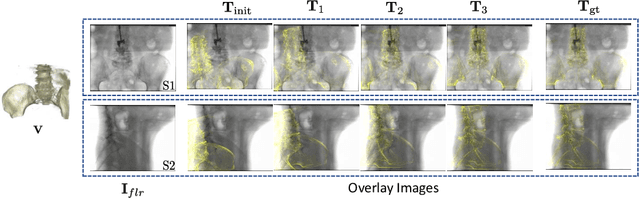
Abstract:Deep Learning-based 2D/3D registration methods are highly robust but often lack the necessary registration accuracy for clinical application. A refinement step using the classical optimization-based 2D/3D registration method applied in combination with Deep Learning-based techniques can provide the required accuracy. However, it also increases the runtime. In this work, we propose a novel Deep Learning driven 2D/3D registration framework that can be used end-to-end for iterative registration tasks without relying on any further refinement step. We accomplish this by learning the update step of the 2D/3D registration framework using Point-to-Plane Correspondences. The update step is learned using iterative residual refinement-based optical flow estimation, in combination with the Point-to-Plane correspondence solver embedded as a known operator. Our proposed method achieves an average runtime of around 8s, a mean re-projection distance error of 0.60 $\pm$ 0.40 mm with a success ratio of 97 percent and a capture range of 60 mm. The combination of high registration accuracy, high robustness, and fast runtime makes our solution ideal for clinical applications.
Deep Learning compatible Differentiable X-ray Projections for Inverse Rendering
Feb 04, 2021

Abstract:Many minimally invasive interventional procedures still rely on 2D fluoroscopic imaging. Generating a patient-specific 3D model from these X-ray projection data would allow to improve the procedural workflow, e.g. by providing assistance functions such as automatic positioning. To accomplish this, two things are required. First, a statistical human shape model of the human anatomy and second, a differentiable X-ray renderer. In this work, we propose a differentiable renderer by deriving the distance travelled by a ray inside mesh structures to generate a distance map. To demonstrate its functioning, we use it for simulating X-ray images from human shape models. Then we show its application by solving the inverse problem, namely reconstructing 3D models from real 2D fluoroscopy images of the pelvis, which is an ideal anatomical structure for patient registration. This is accomplished by an iterative optimization strategy using gradient descent. With the majority of the pelvis being in the fluoroscopic field of view, we achieve a mean Hausdorff distance of 30 mm between the reconstructed model and the ground truth segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge