Elisabeth Hoppe
Pattern Recognition Lab, Department of Computer Science, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany
X-ray Scatter Estimation Using Deep Splines
Jan 22, 2021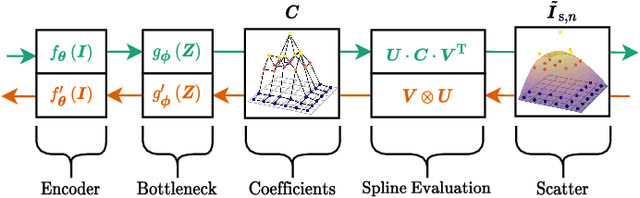
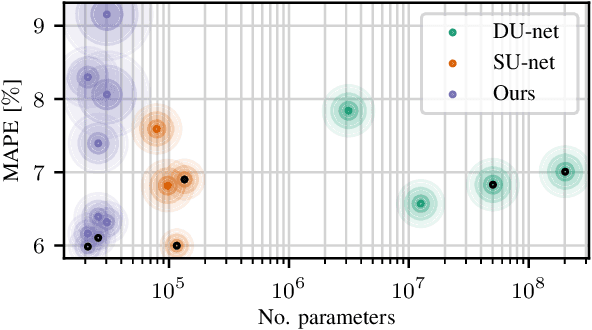
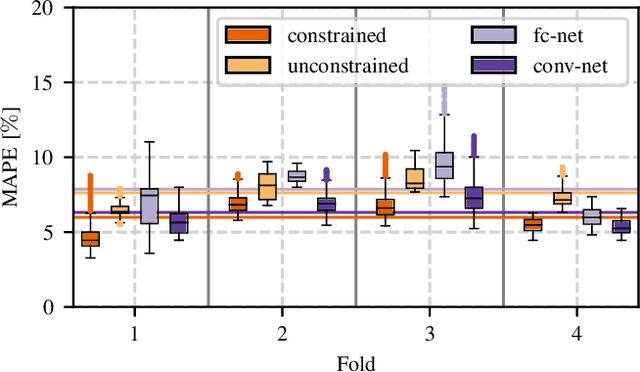
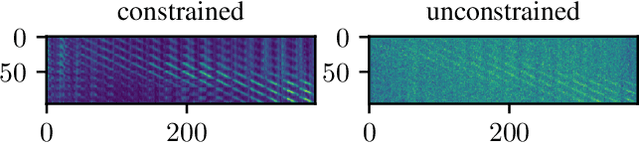
Abstract:Algorithmic X-ray scatter compensation is a desirable technique in flat-panel X-ray imaging and cone-beam computed tomography. State-of-the-art U-net based image translation approaches yielded promising results. As there are no physics constraints applied to the output of the U-Net, it cannot be ruled out that it yields spurious results. Unfortunately, those may be misleading in the context of medical imaging. To overcome this problem, we propose to embed B-splines as a known operator into neural networks. This inherently limits their predictions to well-behaved and smooth functions. In a study using synthetic head and thorax data as well as real thorax phantom data, we found that our approach performed on par with U-net when comparing both algorithms based on quantitative performance metrics. However, our approach not only reduces runtime and parameter complexity, but we also found it much more robust to unseen noise levels. While the U-net responded with visible artifacts, our approach preserved the X-ray signal's frequency characteristics.
2-D Respiration Navigation Framework for 3-D Continuous Cardiac Magnetic Resonance Imaging
Dec 26, 2020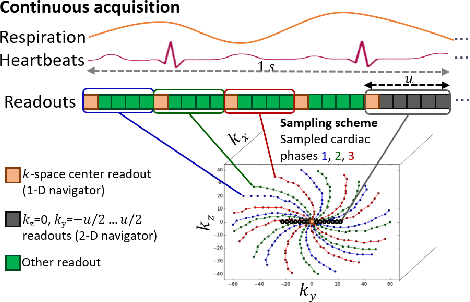
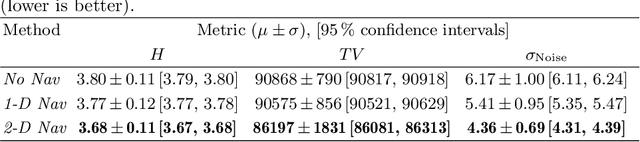
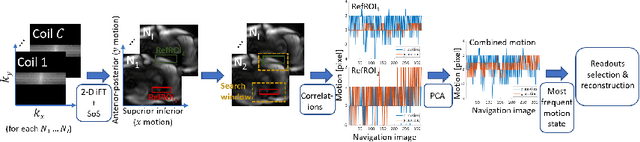
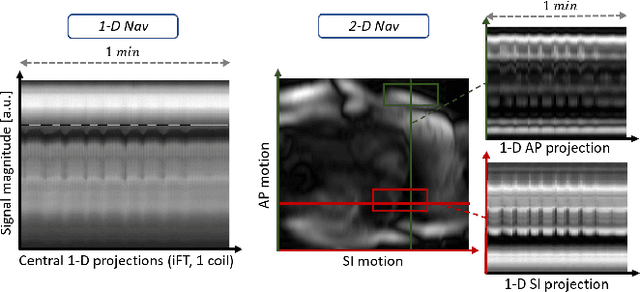
Abstract:Continuous protocols for cardiac magnetic resonance imaging enable sampling of the cardiac anatomy simultaneously resolved into cardiac phases. To avoid respiration artifacts, associated motion during the scan has to be compensated for during reconstruction. In this paper, we propose a sampling adaption to acquire 2-D respiration information during a continuous scan. Further, we develop a pipeline to extract the different respiration states from the acquired signals, which are used to reconstruct data from one respiration phase. Our results show the benefit of the proposed workflow on the image quality compared to no respiration compensation, as well as a previous 1-D respiration navigation approach.
Simultaneous Estimation of X-ray Back-Scatter and Forward-Scatter using Multi-Task Learning
Jul 08, 2020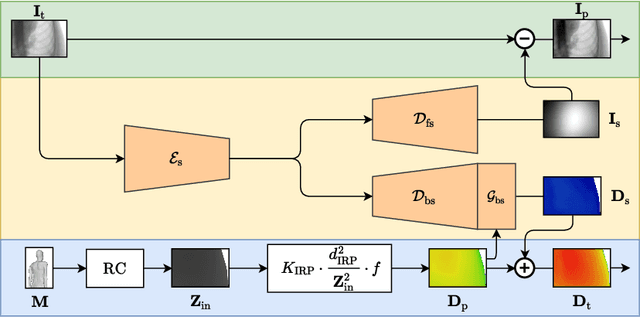
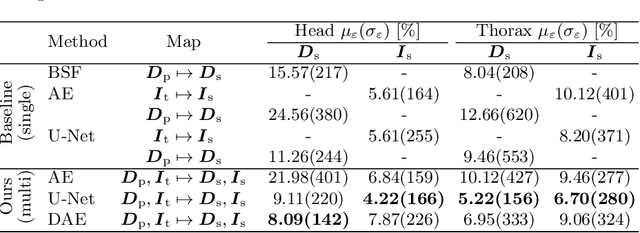
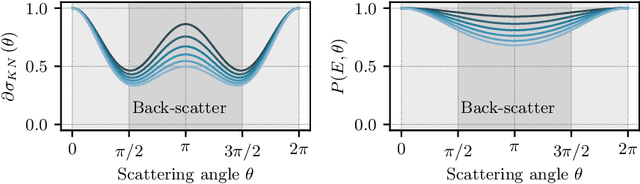
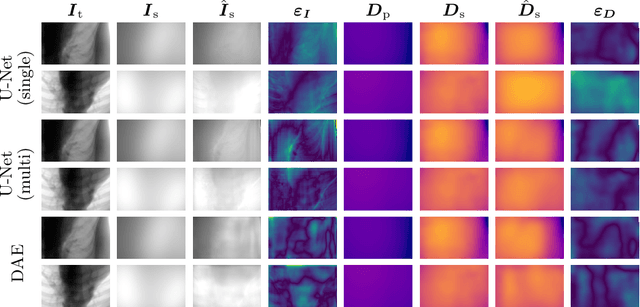
Abstract:Scattered radiation is a major concern impacting X-ray image-guided procedures in two ways. First, back-scatter significantly contributes to patient (skin) dose during complicated interventions. Second, forward-scattered radiation reduces contrast in projection images and introduces artifacts in 3-D reconstructions. While conventionally employed anti-scatter grids improve image quality by blocking X-rays, the additional attenuation due to the anti-scatter grid at the detector needs to be compensated for by a higher patient entrance dose. This also increases the room dose affecting the staff caring for the patient. For skin dose quantification, back-scatter is usually accounted for by applying pre-determined scalar back-scatter factors or linear point spread functions to a primary kerma forward projection onto a patient surface point. However, as patients come in different shapes, the generalization of conventional methods is limited. Here, we propose a novel approach combining conventional techniques with learning-based methods to simultaneously estimate the forward-scatter reaching the detector as well as the back-scatter affecting the patient skin dose. Knowing the forward-scatter, we can correct X-ray projections, while a good estimate of the back-scatter component facilitates an improved skin dose assessment. To simultaneously estimate forward-scatter as well as back-scatter, we propose a multi-task approach for joint back- and forward-scatter estimation by combining X-ray physics with neural networks. We show that, in theory, highly accurate scatter estimation in both cases is possible. In addition, we identify research directions for our multi-task framework and learning-based scatter estimation in general.
Appearance Learning for Image-based Motion Estimation in Tomography
Jun 18, 2020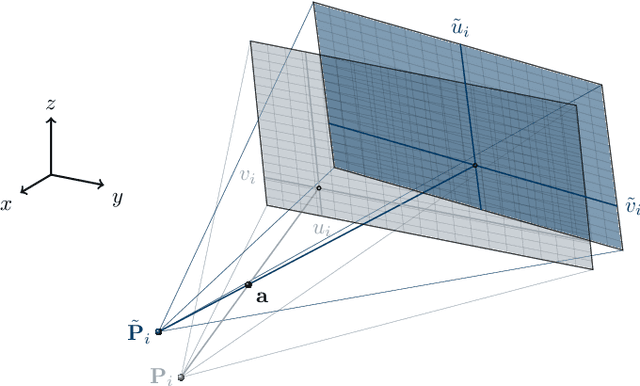
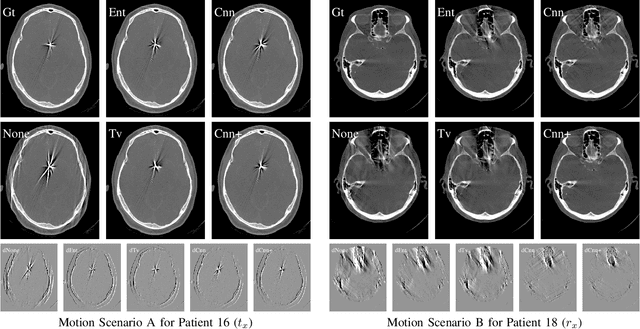
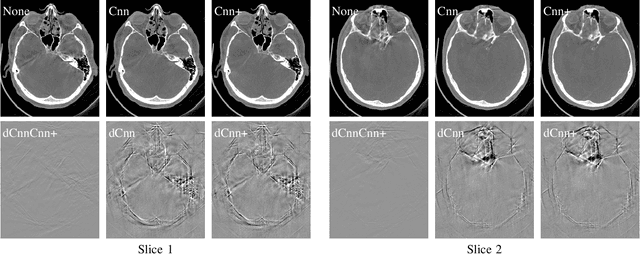
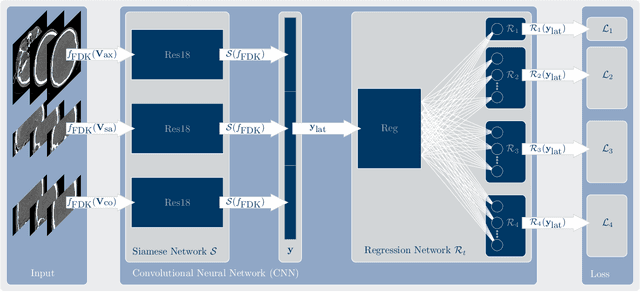
Abstract:In tomographic imaging, anatomical structures are reconstructed by applying a pseudo-inverse forward model to acquired signals. Geometric information within this process is usually depending on the system setting only, i. e., the scanner position or readout direction. Patient motion therefore corrupts the geometry alignment in the reconstruction process resulting in motion artifacts. We propose an appearance learning approach recognizing the structures of rigid motion independently from the scanned object. To this end, we train a siamese triplet network to predict the reprojection error (RPE) for the complete acquisition as well as an approximate distribution of the RPE along the single views from the reconstructed volume in a multi-task learning approach. The RPE measures the motioninduced geometric deviations independent of the object based on virtual marker positions, which are available during training. We train our network using 27 patients and deploy a 21-4-2 split for training, validation and testing. In average, we achieve a residual mean RPE of 0.013mm with an inter-patient standard deviation of 0.022 mm. This is twice the accuracy compared to previously published results. In a motion estimation benchmark the proposed approach achieves superior results in comparison with two state-of-the-art measures in nine out of twelve experiments. The clinical applicability of the proposed method is demonstrated on a motion-affected clinical dataset.
Magnetic Resonance Fingerprinting Reconstruction Using Recurrent Neural Networks
Sep 13, 2019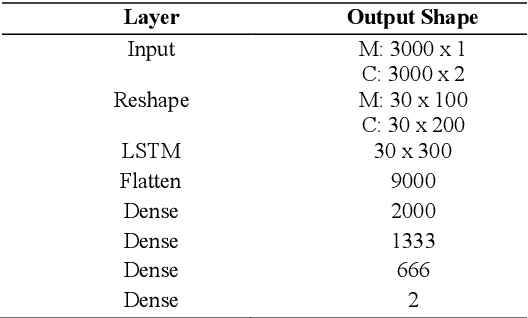
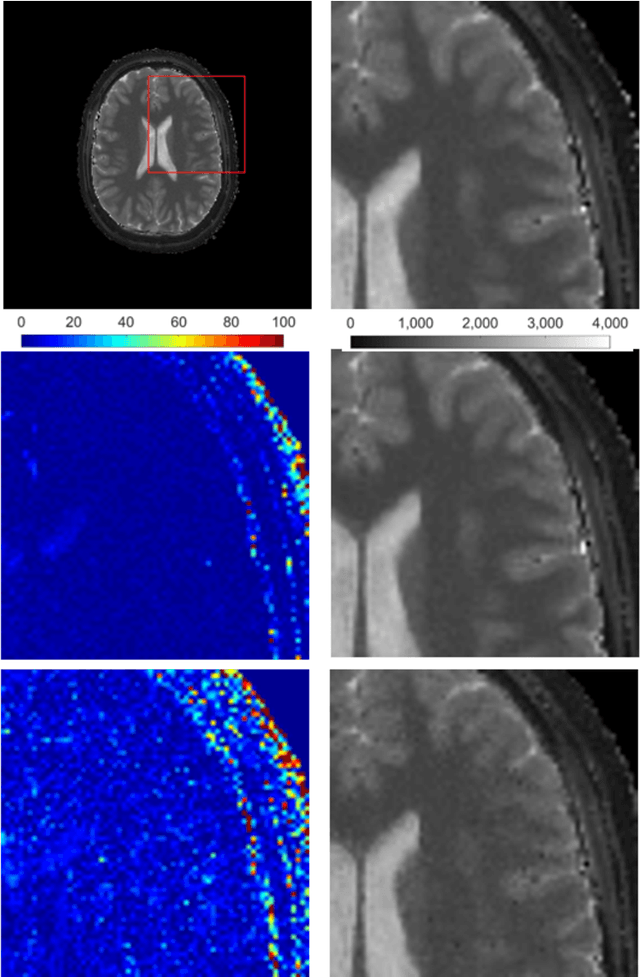
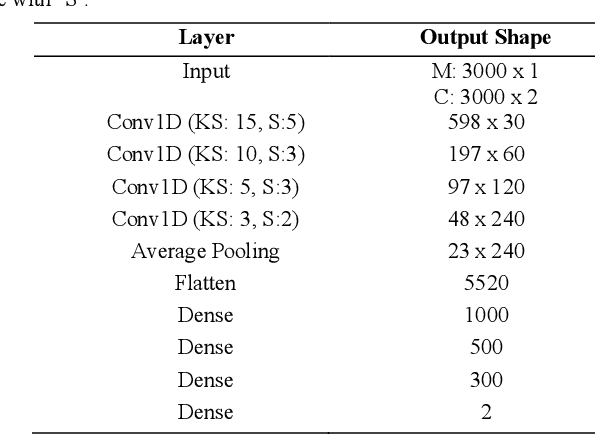
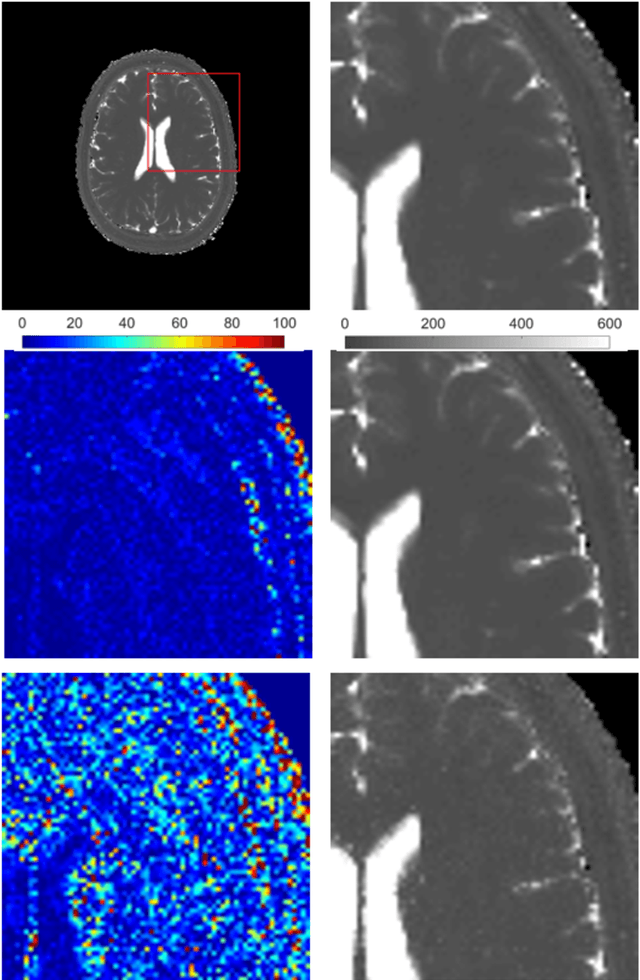
Abstract:Magnetic Resonance Fingerprinting (MRF) is an imaging technique acquiring unique time signals for different tissues. Although the acquisition is highly accelerated, the reconstruction time remains a problem, as the state-of-the-art template matching compares every signal with a set of possible signals. To overcome this limitation, deep learning based approaches, e.g. Convolutional Neural Networks (CNNs) have been proposed. In this work, we investigate the applicability of Recurrent Neural Networks (RNNs) for this reconstruction problem, as the signals are correlated in time. Compared to previous methods based on CNNs, RNN models yield significantly improved results using in-vivo data.
* Accepted and presented at the German Medical Data Sciences (GMDS) conference 2019 (Dortmund, Germany)
RinQ Fingerprinting: Recurrence-informed Quantile Networks for Magnetic Resonance Fingerprinting
Jul 21, 2019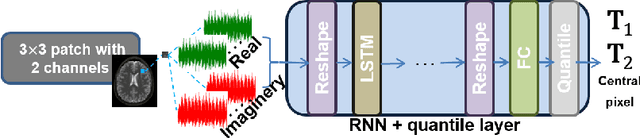
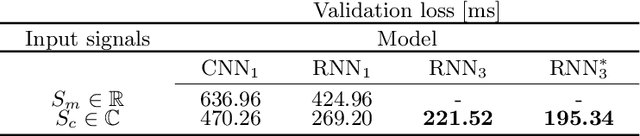
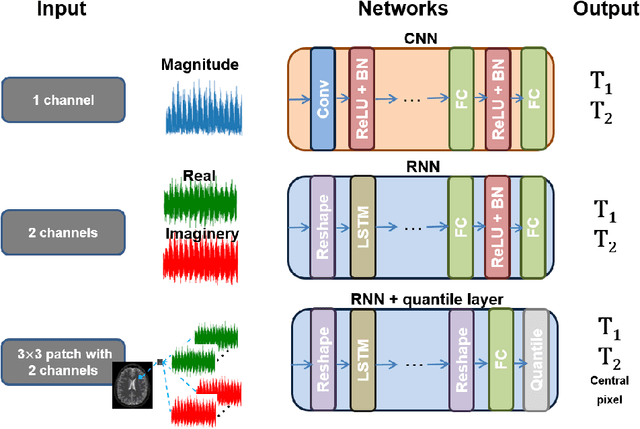
Abstract:Recently, Magnetic Resonance Fingerprinting (MRF) was proposed as a quantitative imaging technique for the simultaneous acquisition of tissue parameters such as relaxation times $T_1$ and $T_2$. Although the acquisition is highly accelerated, the state-of-the-art reconstruction suffers from long computation times: Template matching methods are used to find the most similar signal to the measured one by comparing it to pre-simulated signals of possible parameter combinations in a discretized dictionary. Deep learning approaches can overcome this limitation, by providing the direct mapping from the measured signal to the underlying parameters by one forward pass through a network. In this work, we propose a Recurrent Neural Network (RNN) architecture in combination with a novel quantile layer. RNNs are well suited for the processing of time-dependent signals and the quantile layer helps to overcome the noisy outliers by considering the spatial neighbors of the signal. We evaluate our approach using in-vivo data from multiple brain slices and several volunteers, running various experiments. We show that the RNN approach with small patches of complex-valued input signals in combination with a quantile layer outperforms other architectures, e.g. previously proposed CNNs for the MRF reconstruction reducing the error in $T_1$ and $T_2$ by more than 80%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge