Florian Thamm
Pattern Recognition Lab, Department of Computer Science, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany
Generation of Anonymous Chest Radiographs Using Latent Diffusion Models for Training Thoracic Abnormality Classification Systems
Nov 04, 2022Abstract:The availability of large-scale chest X-ray datasets is a requirement for developing well-performing deep learning-based algorithms in thoracic abnormality detection and classification. However, biometric identifiers in chest radiographs hinder the public sharing of such data for research purposes due to the risk of patient re-identification. To counteract this issue, synthetic data generation offers a solution for anonymizing medical images. This work employs a latent diffusion model to synthesize an anonymous chest X-ray dataset of high-quality class-conditional images. We propose a privacy-enhancing sampling strategy to ensure the non-transference of biometric information during the image generation process. The quality of the generated images and the feasibility of serving as exclusive training data are evaluated on a thoracic abnormality classification task. Compared to a real classifier, we achieve competitive results with a performance gap of only 3.5% in the area under the receiver operating characteristic curve.
Deep Learning-based Anonymization of Chest Radiographs: A Utility-preserving Measure for Patient Privacy
Sep 23, 2022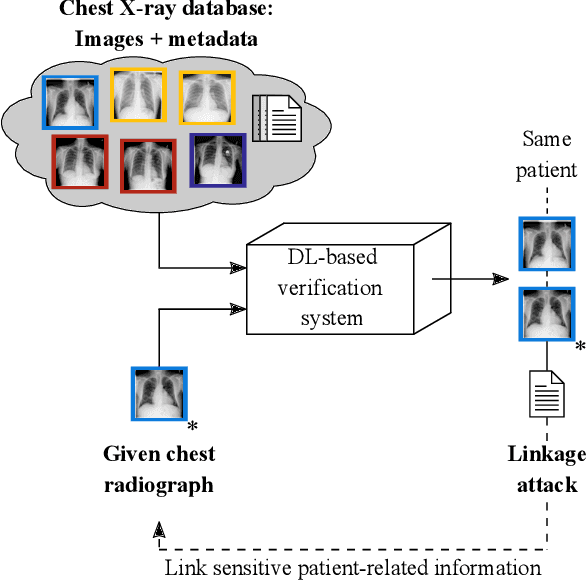
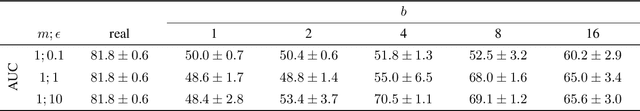
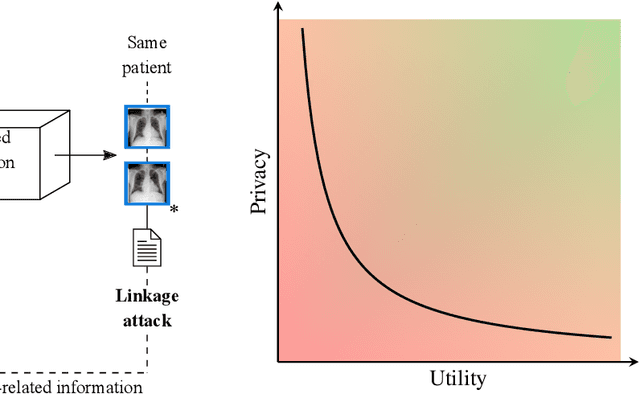
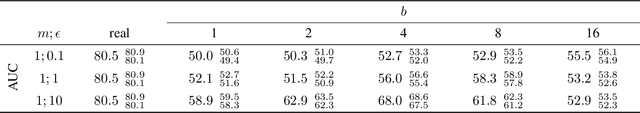
Abstract:Robust and reliable anonymization of chest radiographs constitutes an essential step before publishing large datasets of such for research purposes. The conventional anonymization process is carried out by obscuring personal information in the images with black boxes and removing or replacing meta-information. However, such simple measures retain biometric information in the chest radiographs, allowing patients to be re-identified by a linkage attack. Therefore, we see an urgent need to obfuscate the biometric information appearing in the images. To the best of our knowledge, we propose the first deep learning-based approach to targetedly anonymize chest radiographs while maintaining data utility for diagnostic and machine learning purposes. Our model architecture is a composition of three independent neural networks that, when collectively used, allow for learning a deformation field that is able to impede patient re-identification. The individual influence of each component is investigated with an ablation study. Quantitative results on the ChestX-ray14 dataset show a reduction of patient re-identification from 81.8% to 58.6% in the area under the receiver operating characteristic curve (AUC) with little impact on the abnormality classification performance. This indicates the ability to preserve underlying abnormality patterns while increasing patient privacy. Furthermore, we compare the proposed deep learning-based anonymization approach with differentially private image pixelization, and demonstrate the superiority of our method towards resolving the privacy-utility trade-off for chest radiographs.
Building Brains: Subvolume Recombination for Data Augmentation in Large Vessel Occlusion Detection
May 16, 2022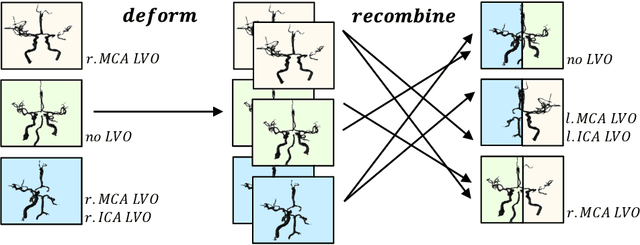
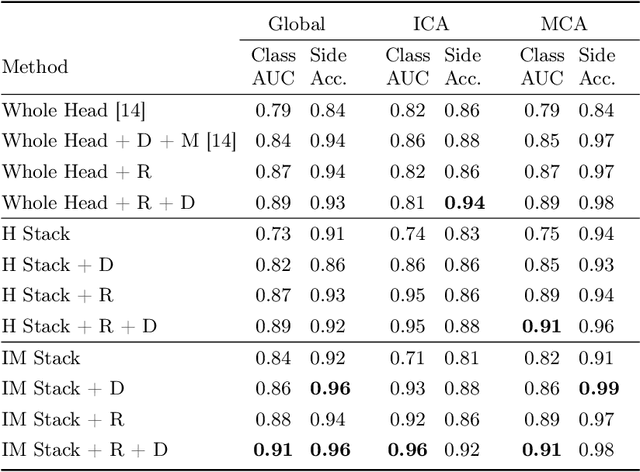
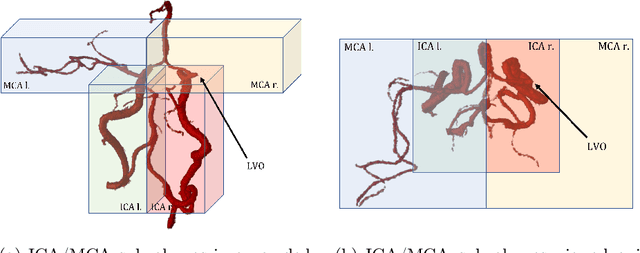
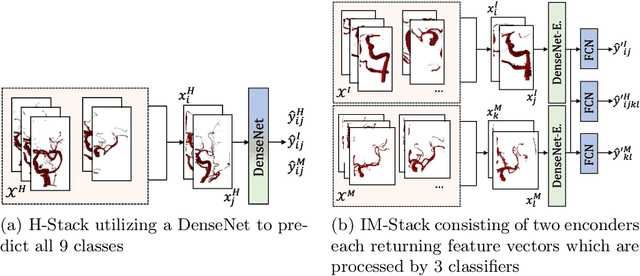
Abstract:Ischemic strokes are often caused by large vessel occlusions (LVOs), which can be visualized and diagnosed with Computed Tomography Angiography scans. As time is brain, a fast, accurate and automated diagnosis of these scans is desirable. Human readers compare the left and right hemispheres in their assessment of strokes. A large training data set is required for a standard deep learning-based model to learn this strategy from data. As labeled medical data in this field is rare, other approaches need to be developed. To both include the prior knowledge of side comparison and increase the amount of training data, we propose an augmentation method that generates artificial training samples by recombining vessel tree segmentations of the hemispheres or hemisphere subregions from different patients. The subregions cover vessels commonly affected by LVOs, namely the internal carotid artery (ICA) and middle cerebral artery (MCA). In line with the augmentation scheme, we use a 3D-DenseNet fed with task-specific input, fostering a side-by-side comparison between the hemispheres. Furthermore, we propose an extension of that architecture to process the individual hemisphere subregions. All configurations predict the presence of an LVO, its side, and the affected subregion. We show the effect of recombination as an augmentation strategy in a 5-fold cross validated ablation study. We enhanced the AUC for patient-wise classification regarding the presence of an LVO of all investigated architectures. For one variant, the proposed method improved the AUC from 0.73 without augmentation to 0.89. The best configuration detects LVOs with an AUC of 0.91, LVOs in the ICA with an AUC of 0.96, and in the MCA with 0.91 while accurately predicting the affected side.
An Algorithm for the Labeling and Interactive Visualization of the Cerebrovascular System of Ischemic Strokes
Apr 26, 2022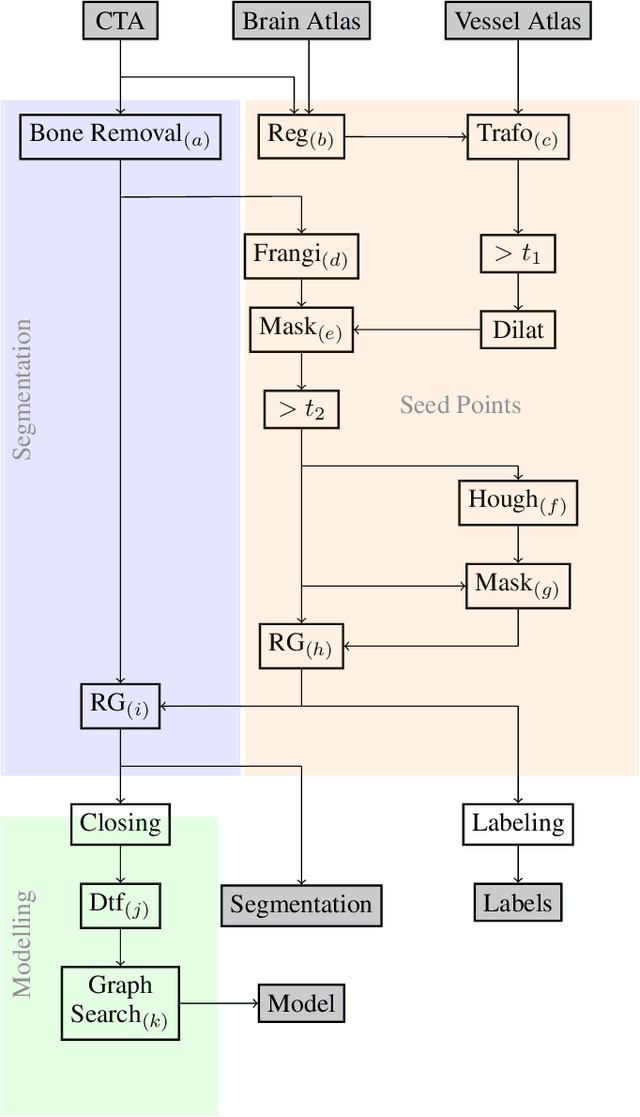
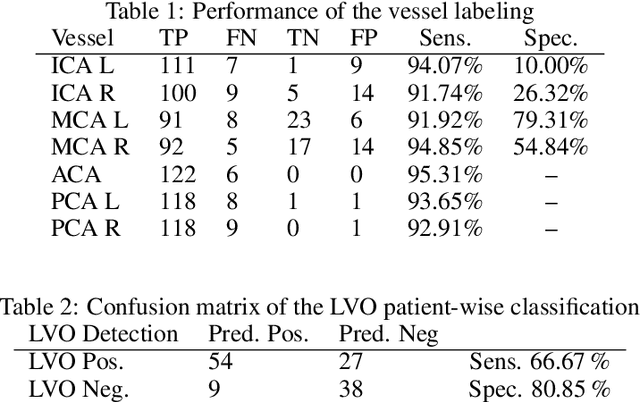
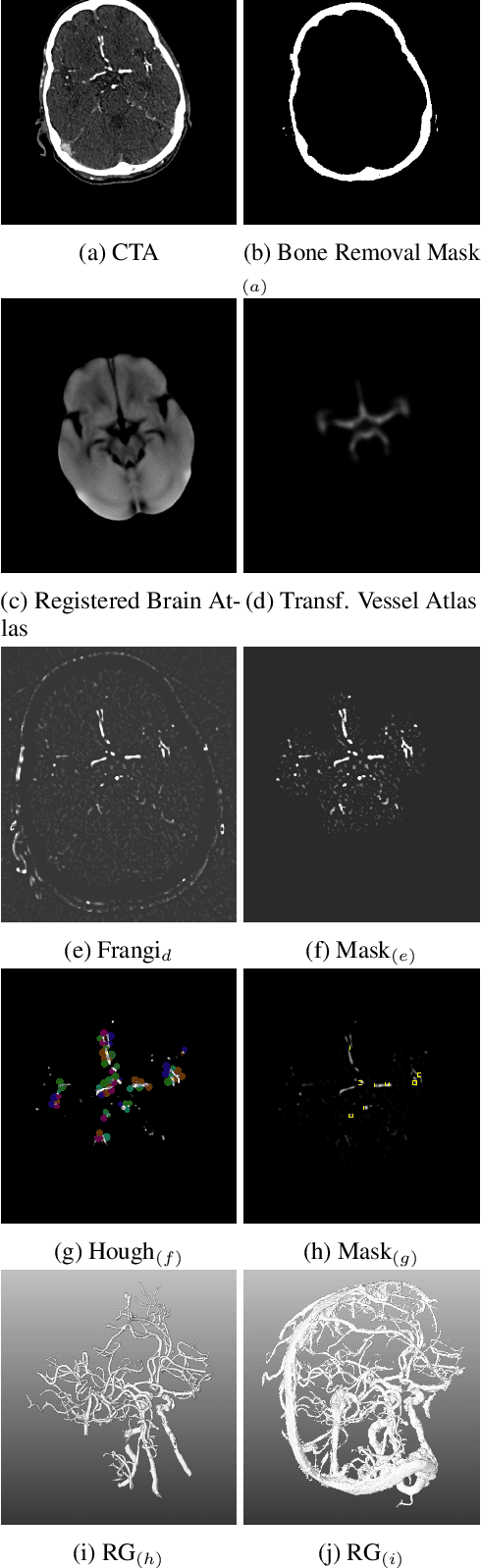

Abstract:During the diagnosis of ischemic strokes, the Circle of Willis and its surrounding vessels are the arteries of interest. Their visualization in case of an acute stroke is often enabled by Computed Tomography Angiography (CTA). Still, the identification and analysis of the cerebral arteries remain time consuming in such scans due to a large number of peripheral vessels which may disturb the visual impression. In previous work we proposed VirtualDSA++, an algorithm designed to segment and label the cerebrovascular tree on CTA scans. Especially with stroke patients, labeling is a delicate procedure, as in the worst case whole hemispheres may not be present due to impeded perfusion. Hence, we extended the labeling mechanism for the cerebral arteries to identify occluded vessels. In the work at hand, we place the algorithm in a clinical context by evaluating the labeling and occlusion detection on stroke patients, where we have achieved labeling sensitivities comparable to other works between 92\,\% and 95\,\%. To the best of our knowledge, ours is the first work to address labeling and occlusion detection at once, whereby a sensitivity of 67\,\% and a specificity of 81\,\% were obtained for the latter. VirtualDSA++ also automatically segments and models the intracranial system, which we further used in a deep learning driven follow up work. We present the generic concept of iterative systematic search for pathways on all nodes of said model, which enables new interactive features. Exemplary, we derive in detail, firstly, the interactive planning of vascular interventions like the mechanical thrombectomy and secondly, the interactive suppression of vessel structures that are not of interest in diagnosing strokes (like veins). We discuss both features as well as further possibilities emerging from the proposed concept.
Segmentation of the Carotid Lumen and Vessel Wall using Deep Learning and Location Priors
Jan 17, 2022
Abstract:In this report we want to present our method and results for the Carotid Artery Vessel Wall Segmentation Challenge. We propose an image-based pipeline utilizing the U-Net architecture and location priors to solve the segmentation problem at hand.
Detection of Large Vessel Occlusions using Deep Learning by Deforming Vessel Tree Segmentations
Dec 10, 2021


Abstract:Computed Tomography Angiography is a key modality providing insights into the cerebrovascular vessel tree that are crucial for the diagnosis and treatment of ischemic strokes, in particular in cases of large vessel occlusions (LVO). Thus, the clinical workflow greatly benefits from an automated detection of patients suffering from LVOs. This work uses convolutional neural networks for case-level classification trained with elastic deformation of the vessel tree segmentation masks to artificially augment training data. Using only masks as the input to our model uniquely allows us to apply such deformations much more aggressively than one could with conventional image volumes while retaining sample realism. The neural network classifies the presence of an LVO and the affected hemisphere. In a 5-fold cross validated ablation study, we demonstrate that the use of the suggested augmentation enables us to train robust models even from few data sets. Training the EfficientNetB1 architecture on 100 data sets, the proposed augmentation scheme was able to raise the ROC AUC to 0.85 from a baseline value of 0.56 using no augmentation. The best performance was achieved using a 3D-DenseNet yielding an AUC of 0.87. The augmentation had positive impact in classification of the affected hemisphere as well, where the 3D-DenseNet reached an AUC of 0.93 on both sides.
Magnetic Resonance Fingerprinting Reconstruction Using Recurrent Neural Networks
Sep 13, 2019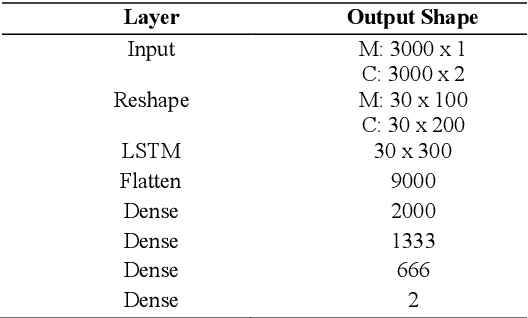
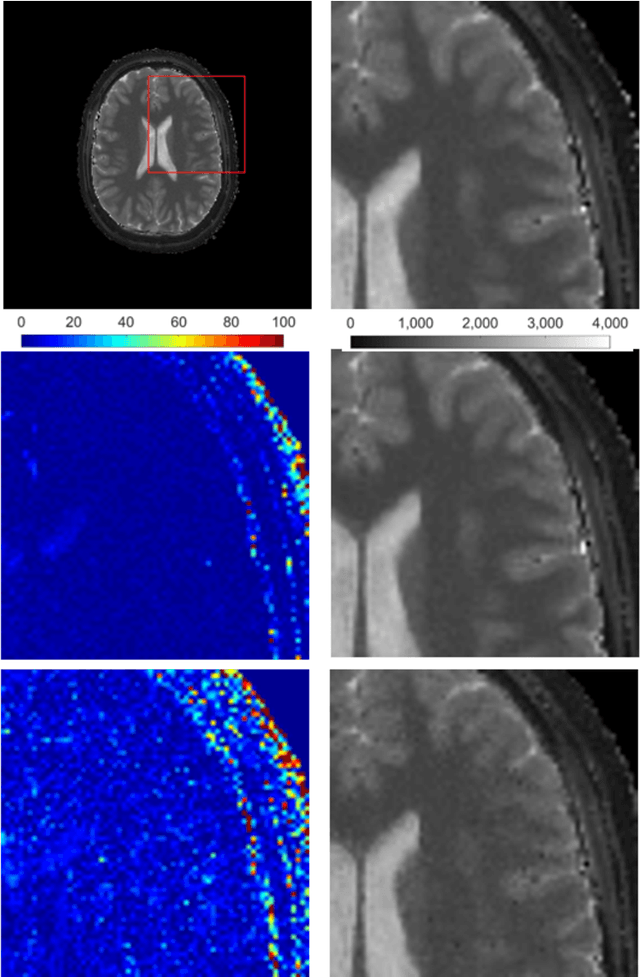
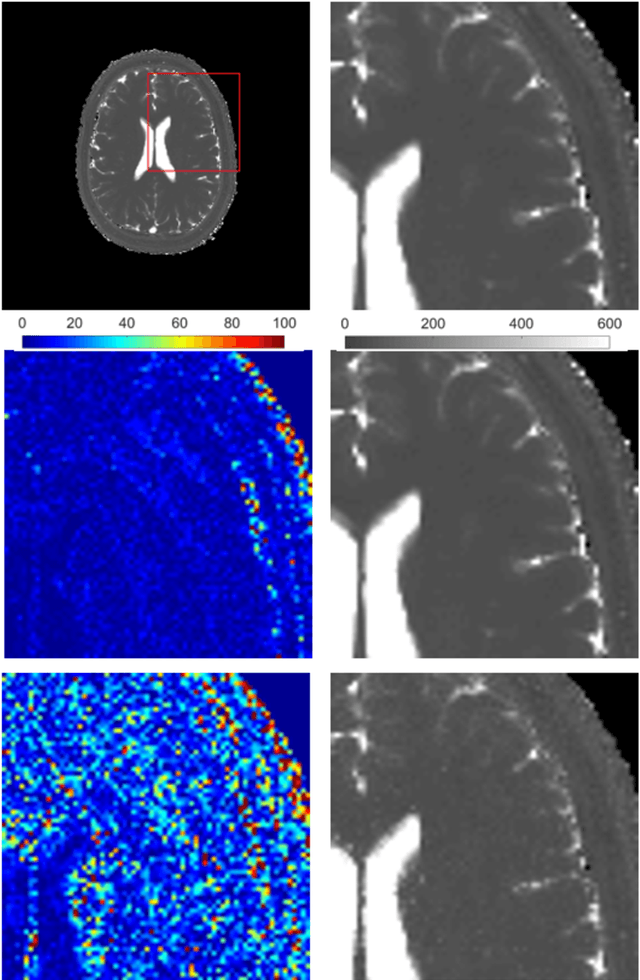
Abstract:Magnetic Resonance Fingerprinting (MRF) is an imaging technique acquiring unique time signals for different tissues. Although the acquisition is highly accelerated, the reconstruction time remains a problem, as the state-of-the-art template matching compares every signal with a set of possible signals. To overcome this limitation, deep learning based approaches, e.g. Convolutional Neural Networks (CNNs) have been proposed. In this work, we investigate the applicability of Recurrent Neural Networks (RNNs) for this reconstruction problem, as the signals are correlated in time. Compared to previous methods based on CNNs, RNN models yield significantly improved results using in-vivo data.
* Accepted and presented at the German Medical Data Sciences (GMDS) conference 2019 (Dortmund, Germany)
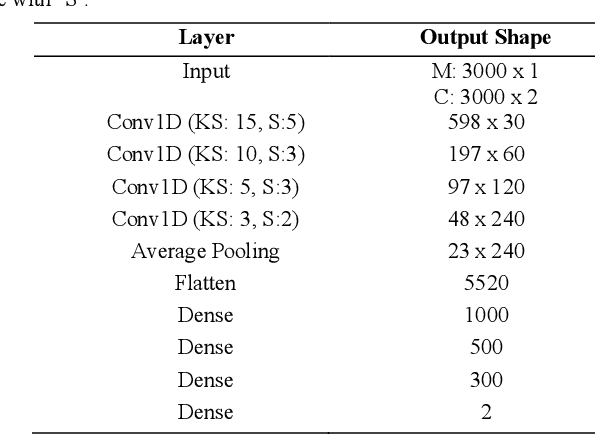
RinQ Fingerprinting: Recurrence-informed Quantile Networks for Magnetic Resonance Fingerprinting
Jul 21, 2019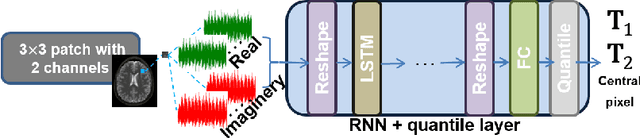
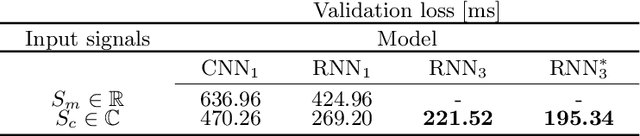
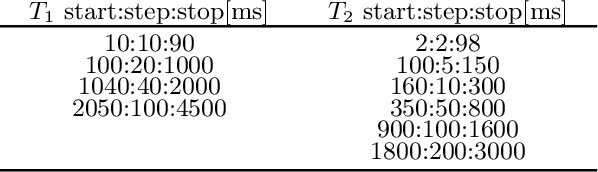
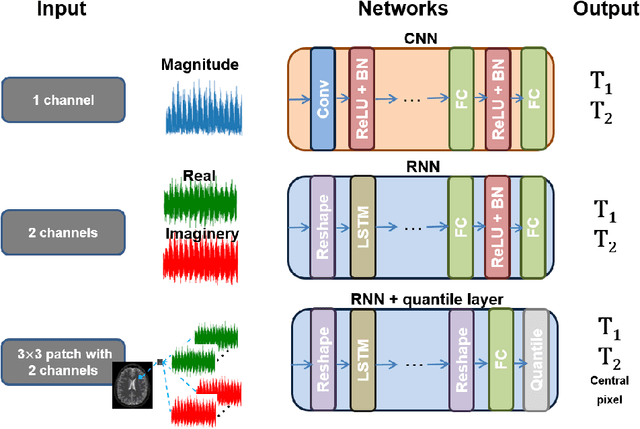
Abstract:Recently, Magnetic Resonance Fingerprinting (MRF) was proposed as a quantitative imaging technique for the simultaneous acquisition of tissue parameters such as relaxation times $T_1$ and $T_2$. Although the acquisition is highly accelerated, the state-of-the-art reconstruction suffers from long computation times: Template matching methods are used to find the most similar signal to the measured one by comparing it to pre-simulated signals of possible parameter combinations in a discretized dictionary. Deep learning approaches can overcome this limitation, by providing the direct mapping from the measured signal to the underlying parameters by one forward pass through a network. In this work, we propose a Recurrent Neural Network (RNN) architecture in combination with a novel quantile layer. RNNs are well suited for the processing of time-dependent signals and the quantile layer helps to overcome the noisy outliers by considering the spatial neighbors of the signal. We evaluate our approach using in-vivo data from multiple brain slices and several volunteers, running various experiments. We show that the RNN approach with small patches of complex-valued input signals in combination with a quantile layer outperforms other architectures, e.g. previously proposed CNNs for the MRF reconstruction reducing the error in $T_1$ and $T_2$ by more than 80%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge