Oliver Taubmann
Siemens Healthcare GmbH, Forchheim, Germany
Neural Image Unfolding: Flattening Sparse Anatomical Structures using Neural Fields
Nov 27, 2024



Abstract:Tomographic imaging reveals internal structures of 3D objects and is crucial for medical diagnoses. Visualizing the morphology and appearance of non-planar sparse anatomical structures that extend over multiple 2D slices in tomographic volumes is inherently difficult but valuable for decision-making and reporting. Hence, various organ-specific unfolding techniques exist to map their densely sampled 3D surfaces to a distortion-minimized 2D representation. However, there is no versatile framework to flatten complex sparse structures including vascular, duct or bone systems. We deploy a neural field to fit the transformation of the anatomy of interest to a 2D overview image. We further propose distortion regularization strategies and combine geometric with intensity-based loss formulations to also display non-annotated and auxiliary targets. In addition to improved versatility, our unfolding technique outperforms mesh-based baselines for sparse structures w.r.t. peak distortion and our regularization scheme yields smoother transformations compared to Jacobian formulations from neural field-based image registration.
SEMPAI: a Self-Enhancing Multi-Photon Artificial Intelligence for prior-informed assessment of muscle function and pathology
Oct 28, 2022Abstract:Deep learning (DL) shows notable success in biomedical studies. However, most DL algorithms work as a black box, exclude biomedical experts, and need extensive data. We introduce the Self-Enhancing Multi-Photon Artificial Intelligence (SEMPAI), that integrates hypothesis-driven priors in a data-driven DL approach for research on multiphoton microscopy (MPM) of muscle fibers. SEMPAI utilizes meta-learning to optimize prior integration, data representation, and neural network architecture simultaneously. This allows hypothesis testing and provides interpretable feedback about the origin of biological information in MPM images. SEMPAI performs joint learning of several tasks to enable prediction for small datasets. The method is applied on an extensive multi-study dataset resulting in the largest joint analysis of pathologies and function for single muscle fibers. SEMPAI outperforms state-of-the-art biomarkers in six of seven predictive tasks, including those with scarce data. SEMPAI's DL models with integrated priors are superior to those without priors and to prior-only machine learning approaches.
DeepTechnome: Mitigating Unknown Bias in Deep Learning Based Assessment of CT Images
May 26, 2022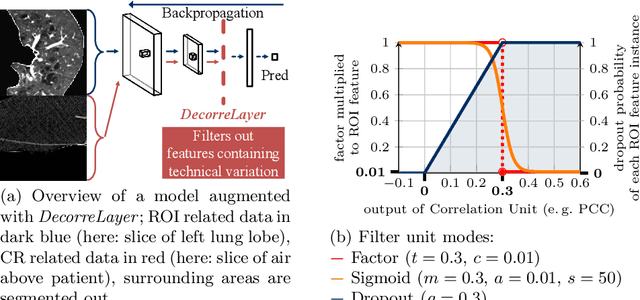
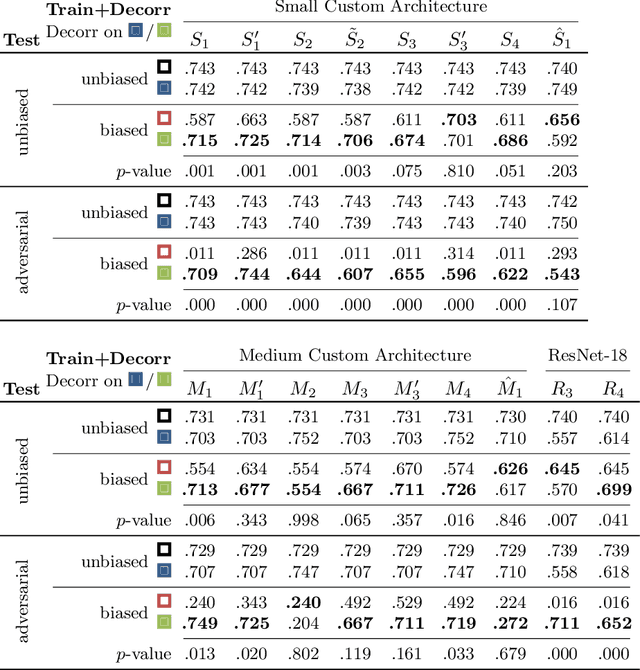

Abstract:Reliably detecting diseases using relevant biological information is crucial for real-world applicability of deep learning techniques in medical imaging. We debias deep learning models during training against unknown bias - without preprocessing/filtering the input beforehand or assuming specific knowledge about its distribution or precise nature in the dataset. We use control regions as surrogates that carry information regarding the bias, employ the classifier model to extract features, and suppress biased intermediate features with our custom, modular DecorreLayer. We evaluate our method on a dataset of 952 lung computed tomography scans by introducing simulated biases w.r.t. reconstruction kernel and noise level and propose including an adversarial test set in evaluations of bias reduction techniques. In a moderately sized model architecture, applying the proposed method to learn from data exhibiting a strong bias, it near-perfectly recovers the classification performance observed when training with corresponding unbiased data.
Building Brains: Subvolume Recombination for Data Augmentation in Large Vessel Occlusion Detection
May 16, 2022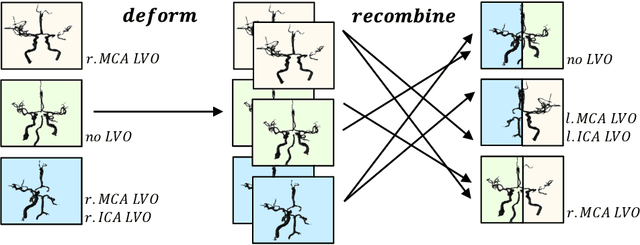
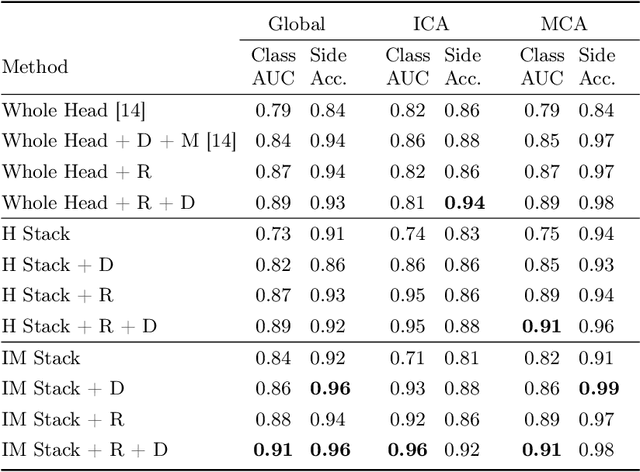
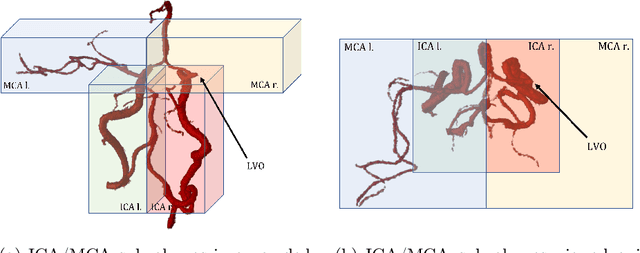
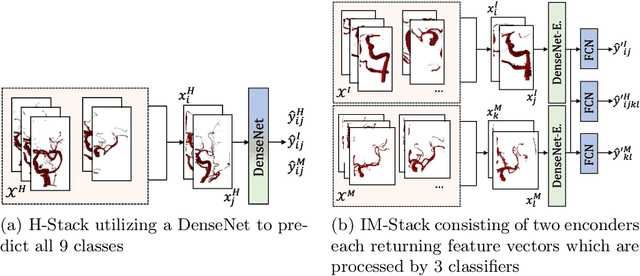
Abstract:Ischemic strokes are often caused by large vessel occlusions (LVOs), which can be visualized and diagnosed with Computed Tomography Angiography scans. As time is brain, a fast, accurate and automated diagnosis of these scans is desirable. Human readers compare the left and right hemispheres in their assessment of strokes. A large training data set is required for a standard deep learning-based model to learn this strategy from data. As labeled medical data in this field is rare, other approaches need to be developed. To both include the prior knowledge of side comparison and increase the amount of training data, we propose an augmentation method that generates artificial training samples by recombining vessel tree segmentations of the hemispheres or hemisphere subregions from different patients. The subregions cover vessels commonly affected by LVOs, namely the internal carotid artery (ICA) and middle cerebral artery (MCA). In line with the augmentation scheme, we use a 3D-DenseNet fed with task-specific input, fostering a side-by-side comparison between the hemispheres. Furthermore, we propose an extension of that architecture to process the individual hemisphere subregions. All configurations predict the presence of an LVO, its side, and the affected subregion. We show the effect of recombination as an augmentation strategy in a 5-fold cross validated ablation study. We enhanced the AUC for patient-wise classification regarding the presence of an LVO of all investigated architectures. For one variant, the proposed method improved the AUC from 0.73 without augmentation to 0.89. The best configuration detects LVOs with an AUC of 0.91, LVOs in the ICA with an AUC of 0.96, and in the MCA with 0.91 while accurately predicting the affected side.
An Algorithm for the Labeling and Interactive Visualization of the Cerebrovascular System of Ischemic Strokes
Apr 26, 2022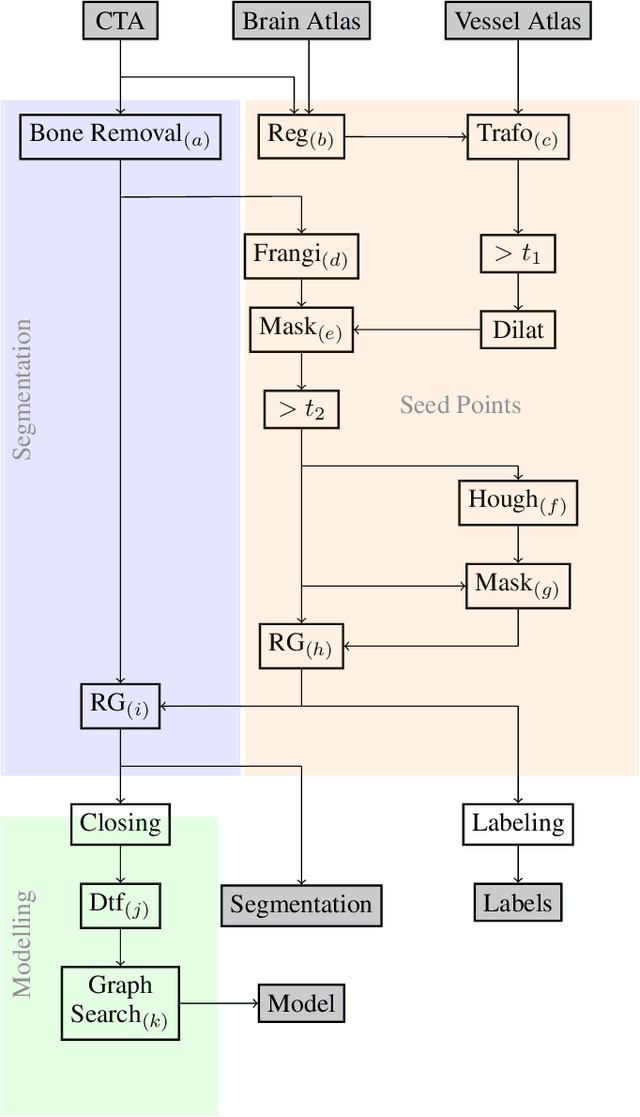
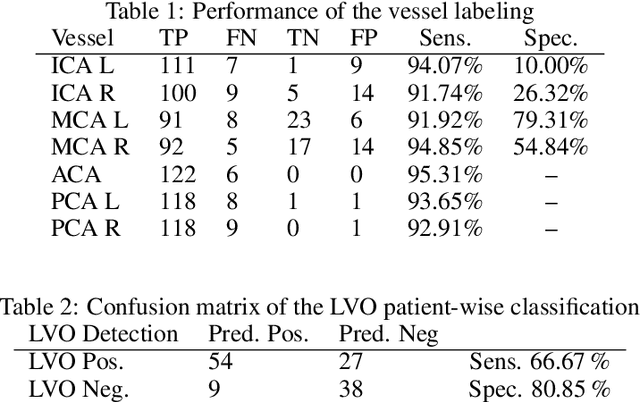
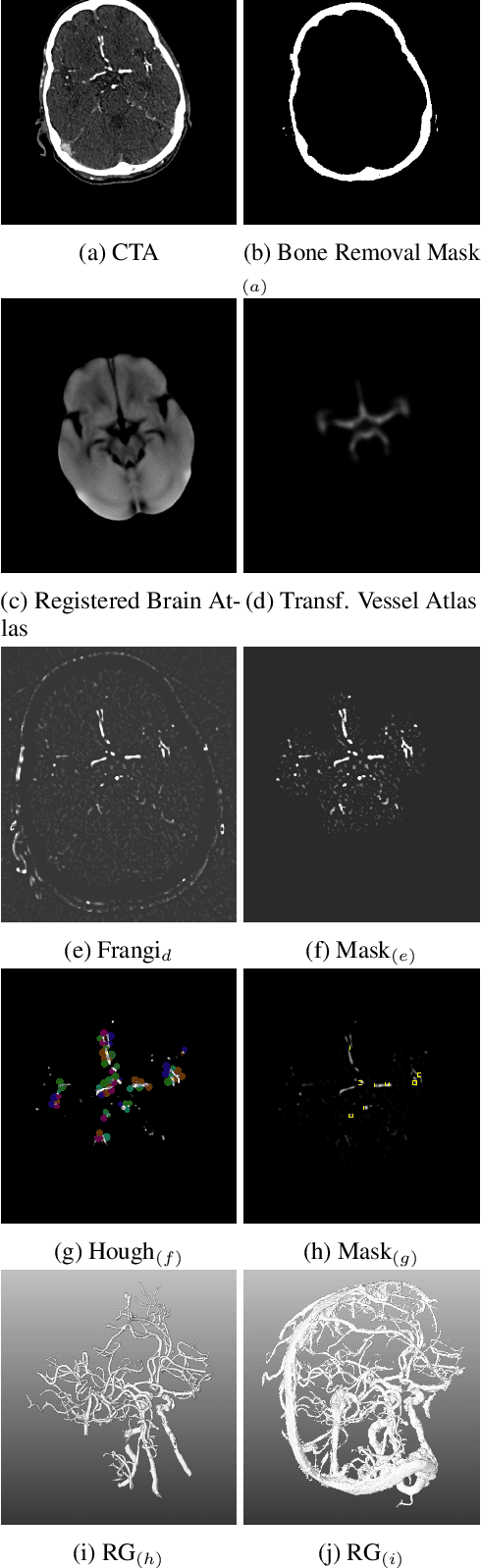

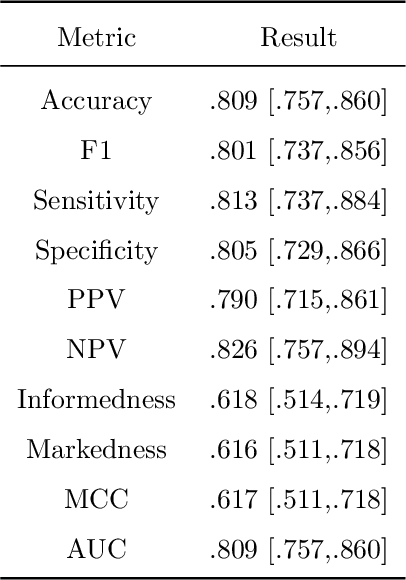
Abstract:During the diagnosis of ischemic strokes, the Circle of Willis and its surrounding vessels are the arteries of interest. Their visualization in case of an acute stroke is often enabled by Computed Tomography Angiography (CTA). Still, the identification and analysis of the cerebral arteries remain time consuming in such scans due to a large number of peripheral vessels which may disturb the visual impression. In previous work we proposed VirtualDSA++, an algorithm designed to segment and label the cerebrovascular tree on CTA scans. Especially with stroke patients, labeling is a delicate procedure, as in the worst case whole hemispheres may not be present due to impeded perfusion. Hence, we extended the labeling mechanism for the cerebral arteries to identify occluded vessels. In the work at hand, we place the algorithm in a clinical context by evaluating the labeling and occlusion detection on stroke patients, where we have achieved labeling sensitivities comparable to other works between 92\,\% and 95\,\%. To the best of our knowledge, ours is the first work to address labeling and occlusion detection at once, whereby a sensitivity of 67\,\% and a specificity of 81\,\% were obtained for the latter. VirtualDSA++ also automatically segments and models the intracranial system, which we further used in a deep learning driven follow up work. We present the generic concept of iterative systematic search for pathways on all nodes of said model, which enables new interactive features. Exemplary, we derive in detail, firstly, the interactive planning of vascular interventions like the mechanical thrombectomy and secondly, the interactive suppression of vessel structures that are not of interest in diagnosing strokes (like veins). We discuss both features as well as further possibilities emerging from the proposed concept.
CAD-RADS Scoring using Deep Learning and Task-Specific Centerline Labeling
Feb 08, 2022



Abstract:With coronary artery disease (CAD) persisting to be one of the leading causes of death worldwide, interest in supporting physicians with algorithms to speed up and improve diagnosis is high. In clinical practice, the severeness of CAD is often assessed with a coronary CT angiography (CCTA) scan and manually graded with the CAD-Reporting and Data System (CAD-RADS) score. The clinical questions this score assesses are whether patients have CAD or not (rule-out) and whether they have severe CAD or not (hold-out). In this work, we reach new state-of-the-art performance for automatic CAD-RADS scoring. We propose using severity-based label encoding, test time augmentation (TTA) and model ensembling for a task-specific deep learning architecture. Furthermore, we introduce a novel task- and model-specific, heuristic coronary segment labeling, which subdivides coronary trees into consistent parts across patients. It is fast, robust, and easy to implement. We were able to raise the previously reported area under the receiver operating characteristic curve (AUC) from 0.914 to 0.942 in the rule-out and from 0.921 to 0.950 in the hold-out task respectively.
Detection of Large Vessel Occlusions using Deep Learning by Deforming Vessel Tree Segmentations
Dec 10, 2021


Abstract:Computed Tomography Angiography is a key modality providing insights into the cerebrovascular vessel tree that are crucial for the diagnosis and treatment of ischemic strokes, in particular in cases of large vessel occlusions (LVO). Thus, the clinical workflow greatly benefits from an automated detection of patients suffering from LVOs. This work uses convolutional neural networks for case-level classification trained with elastic deformation of the vessel tree segmentation masks to artificially augment training data. Using only masks as the input to our model uniquely allows us to apply such deformations much more aggressively than one could with conventional image volumes while retaining sample realism. The neural network classifies the presence of an LVO and the affected hemisphere. In a 5-fold cross validated ablation study, we demonstrate that the use of the suggested augmentation enables us to train robust models even from few data sets. Training the EfficientNetB1 architecture on 100 data sets, the proposed augmentation scheme was able to raise the ROC AUC to 0.85 from a baseline value of 0.56 using no augmentation. The best performance was achieved using a 3D-DenseNet yielding an AUC of 0.87. The augmentation had positive impact in classification of the affected hemisphere as well, where the 3D-DenseNet reached an AUC of 0.93 on both sides.
Explaining Clinical Decision Support Systems in Medical Imaging using Cycle-Consistent Activation Maximization
Oct 13, 2020
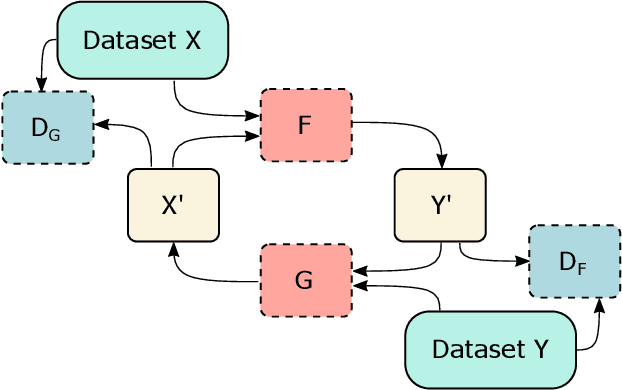
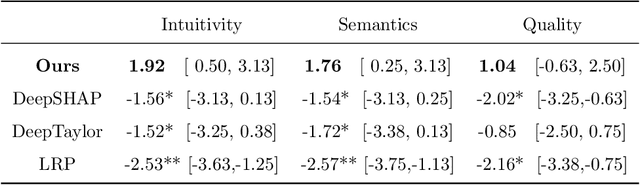
Abstract:Clinical decision support using deep neural networks has become a topic of steadily growing interest. While recent work has repeatedly demonstrated that deep learning offers major advantages for medical image classification over traditional methods, clinicians are often hesitant to adopt the technology because its underlying decision-making process is considered to be intransparent and difficult to comprehend. In recent years, this has been addressed by a variety of approaches that have successfully contributed to providing deeper insight. Most notably, additive feature attribution methods are able to propagate decisions back into the input space by creating a saliency map which allows the practitioner to "see what the network sees." However, the quality of the generated maps can become poor and the images noisy if only limited data is available - a typical scenario in clinical contexts. We propose a novel decision explanation scheme based on CycleGAN activation maximization which generates high-quality visualizations of classifier decisions even in smaller data sets. We conducted a user study in which these visualizations significantly outperformed existing methods on the LIDC dataset for lung lesion malignancy classification. With our approach we make a significant contribution to a better understanding of clinical decision support systems based on deep neural networks and thus aim to foster overall clinical acceptance.
Scale-Space Anisotropic Total Variation for Limited Angle Tomography
Jan 29, 2018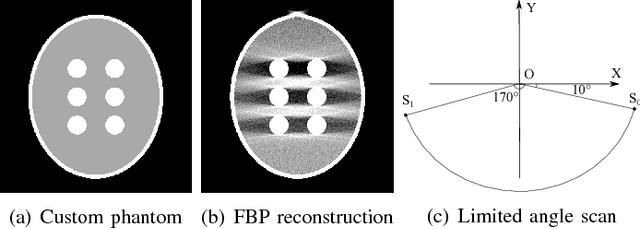

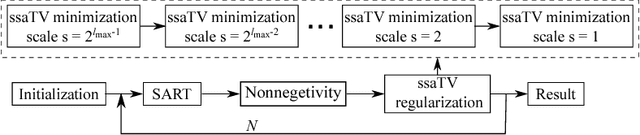

Abstract:This paper addresses streak reduction in limited angle tomography. Although the iterative reweighted total variation (wTV) algorithm reduces small streaks well, it is rather inept at eliminating large ones since total variation (TV) regularization is scale-dependent and may regard these streaks as homogeneous areas. Hence, the main purpose of this paper is to reduce streak artifacts at various scales. We propose the scale-space anisotropic total variation (ssaTV) algorithm in two different implementations. The first implementation (ssaTV-1) utilizes an anisotropic gradient-like operator which uses 2s neighboring pixels along the streaks' normal direction at each scale s. The second implementation (ssaTV-2) makes use of anisotropic down-sampling and up-sampling operations, similarly oriented along the streaks' normal direction, to apply TV regularization at various scales. Experiments on numerical and clinical data demonstrate that both ssaTV algorithms reduce streak artifacts more effectively and efficiently than wTV, particularly when using multiple scales.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge