Felix Denzinger
Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Germany, Siemens Healthcare GmbH, Forchheim, Germany
Gradient-Based Geometry Learning for Fan-Beam CT Reconstruction
Dec 05, 2022Abstract:Incorporating computed tomography (CT) reconstruction operators into differentiable pipelines has proven beneficial in many applications. Such approaches usually focus on the projection data and keep the acquisition geometry fixed. However, precise knowledge of the acquisition geometry is essential for high quality reconstruction results. In this paper, the differentiable formulation of fan-beam CT reconstruction is extended to the acquisition geometry. This allows to propagate gradient information from a loss function on the reconstructed image into the geometry parameters. As a proof-of-concept experiment, this idea is applied to rigid motion compensation. The cost function is parameterized by a trained neural network which regresses an image quality metric from the motion affected reconstruction alone. Using the proposed method, we are the first to optimize such an autofocus-inspired algorithm based on analytical gradients. The algorithm achieves a reduction in MSE by 35.5 % and an improvement in SSIM by 12.6 % over the motion affected reconstruction. Next to motion compensation, we see further use cases of our differentiable method for scanner calibration or hybrid techniques employing deep models.
An unobtrusive quality supervision approach for medical image annotation
Nov 22, 2022Abstract:Image annotation is one essential prior step to enable data-driven algorithms. In medical imaging, having large and reliably annotated data sets is crucial to recognize various diseases robustly. However, annotator performance varies immensely, thus impacts model training. Therefore, often multiple annotators should be employed, which is however expensive and resource-intensive. Hence, it is desirable that users should annotate unseen data and have an automated system to unobtrusively rate their performance during this process. We examine such a system based on whole slide images (WSIs) showing lung fluid cells. We evaluate two methods the generation of synthetic individual cell images: conditional Generative Adversarial Networks and Diffusion Models (DM). For qualitative and quantitative evaluation, we conduct a user study to highlight the suitability of generated cells. Users could not detect 52.12% of generated images by DM proofing the feasibility to replace the original cells with synthetic cells without being noticed.
On the Benefit of Dual-domain Denoising in a Self-supervised Low-dose CT Setting
Nov 03, 2022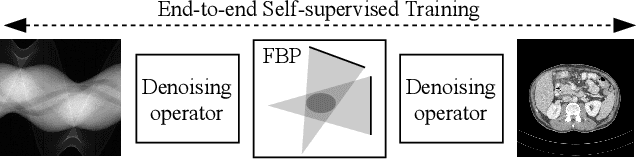
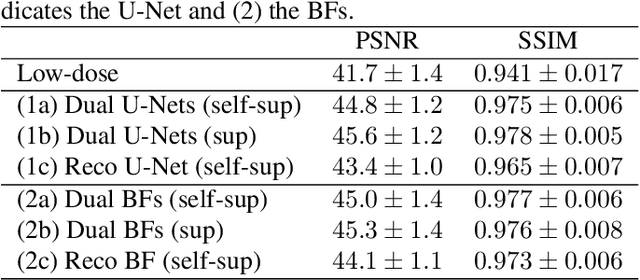
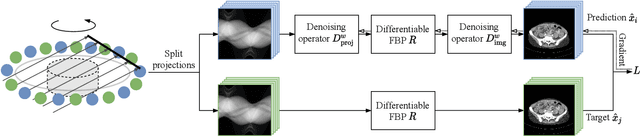
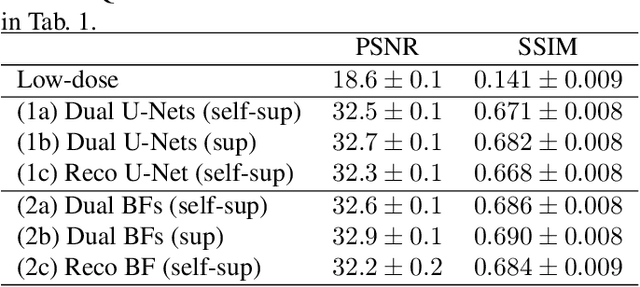
Abstract:Computed tomography (CT) is routinely used for three-dimensional non-invasive imaging. Numerous data-driven image denoising algorithms were proposed to restore image quality in low-dose acquisitions. However, considerably less research investigates methods already intervening in the raw detector data due to limited access to suitable projection data or correct reconstruction algorithms. In this work, we present an end-to-end trainable CT reconstruction pipeline that contains denoising operators in both the projection and the image domain and that are optimized simultaneously without requiring ground-truth high-dose CT data. Our experiments demonstrate that including an additional projection denoising operator improved the overall denoising performance by 82.4-94.1%/12.5-41.7% (PSNR/SSIM) on abdomen CT and 1.5-2.9%/0.4-0.5% (PSNR/SSIM) on XRM data relative to the low-dose baseline. We make our entire helical CT reconstruction framework publicly available that contains a raw projection rebinning step to render helical projection data suitable for differentiable fan-beam reconstruction operators and end-to-end learning.
SEMPAI: a Self-Enhancing Multi-Photon Artificial Intelligence for prior-informed assessment of muscle function and pathology
Oct 28, 2022Abstract:Deep learning (DL) shows notable success in biomedical studies. However, most DL algorithms work as a black box, exclude biomedical experts, and need extensive data. We introduce the Self-Enhancing Multi-Photon Artificial Intelligence (SEMPAI), that integrates hypothesis-driven priors in a data-driven DL approach for research on multiphoton microscopy (MPM) of muscle fibers. SEMPAI utilizes meta-learning to optimize prior integration, data representation, and neural network architecture simultaneously. This allows hypothesis testing and provides interpretable feedback about the origin of biological information in MPM images. SEMPAI performs joint learning of several tasks to enable prediction for small datasets. The method is applied on an extensive multi-study dataset resulting in the largest joint analysis of pathologies and function for single muscle fibers. SEMPAI outperforms state-of-the-art biomarkers in six of seven predictive tasks, including those with scarce data. SEMPAI's DL models with integrated priors are superior to those without priors and to prior-only machine learning approaches.
Deep Learning-based Anonymization of Chest Radiographs: A Utility-preserving Measure for Patient Privacy
Sep 23, 2022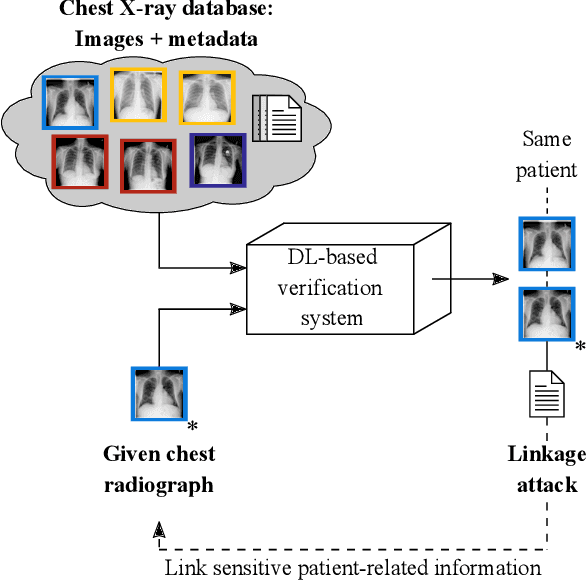
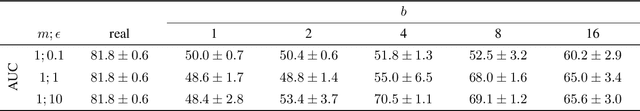
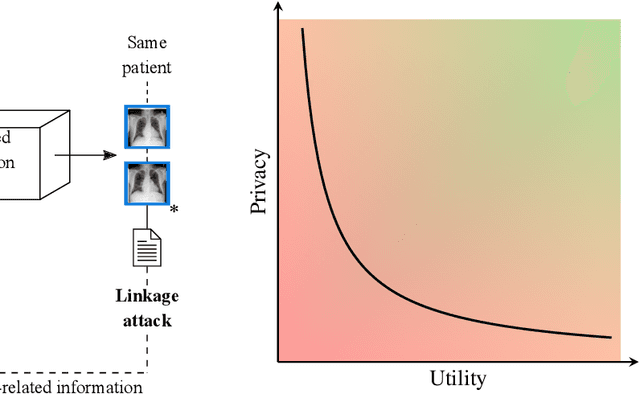
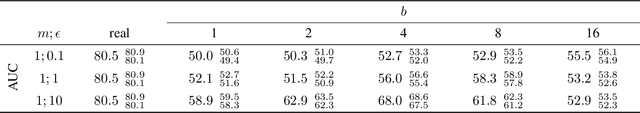
Abstract:Robust and reliable anonymization of chest radiographs constitutes an essential step before publishing large datasets of such for research purposes. The conventional anonymization process is carried out by obscuring personal information in the images with black boxes and removing or replacing meta-information. However, such simple measures retain biometric information in the chest radiographs, allowing patients to be re-identified by a linkage attack. Therefore, we see an urgent need to obfuscate the biometric information appearing in the images. To the best of our knowledge, we propose the first deep learning-based approach to targetedly anonymize chest radiographs while maintaining data utility for diagnostic and machine learning purposes. Our model architecture is a composition of three independent neural networks that, when collectively used, allow for learning a deformation field that is able to impede patient re-identification. The individual influence of each component is investigated with an ablation study. Quantitative results on the ChestX-ray14 dataset show a reduction of patient re-identification from 81.8% to 58.6% in the area under the receiver operating characteristic curve (AUC) with little impact on the abnormality classification performance. This indicates the ability to preserve underlying abnormality patterns while increasing patient privacy. Furthermore, we compare the proposed deep learning-based anonymization approach with differentially private image pixelization, and demonstrate the superiority of our method towards resolving the privacy-utility trade-off for chest radiographs.
Trainable Joint Bilateral Filters for Enhanced Prediction Stability in Low-dose CT
Jul 15, 2022



Abstract:Low-dose computed tomography (CT) denoising algorithms aim to enable reduced patient dose in routine CT acquisitions while maintaining high image quality. Recently, deep learning~(DL)-based methods were introduced, outperforming conventional denoising algorithms on this task due to their high model capacity. However, for the transition of DL-based denoising to clinical practice, these data-driven approaches must generalize robustly beyond the seen training data. We, therefore, propose a hybrid denoising approach consisting of a set of trainable joint bilateral filters (JBFs) combined with a convolutional DL-based denoising network to predict the guidance image. Our proposed denoising pipeline combines the high model capacity enabled by DL-based feature extraction with the reliability of the conventional JBF. The pipeline's ability to generalize is demonstrated by training on abdomen CT scans without metal implants and testing on abdomen scans with metal implants as well as on head CT data. When embedding two well-established DL-based denoisers (RED-CNN/QAE) in our pipeline, the denoising performance is improved by $10\,\%$/$82\,\%$ (RMSE) and $3\,\%$/$81\,\%$ (PSNR) in regions containing metal and by $6\,\%$/$78\,\%$ (RMSE) and $2\,\%$/$4\,\%$ (PSNR) on head CT data, compared to the respective vanilla model. Concluding, the proposed trainable JBFs limit the error bound of deep neural networks to facilitate the applicability of DL-based denoisers in low-dose CT pipelines.
DeepTechnome: Mitigating Unknown Bias in Deep Learning Based Assessment of CT Images
May 26, 2022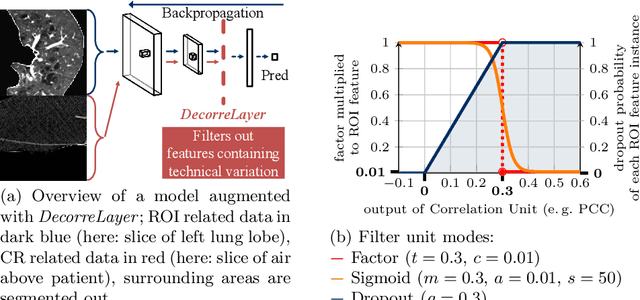
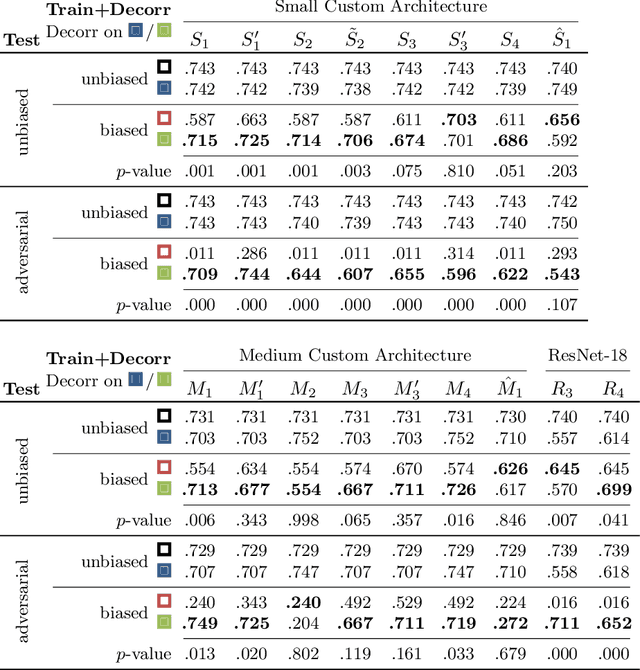

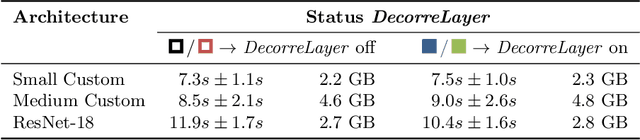
Abstract:Reliably detecting diseases using relevant biological information is crucial for real-world applicability of deep learning techniques in medical imaging. We debias deep learning models during training against unknown bias - without preprocessing/filtering the input beforehand or assuming specific knowledge about its distribution or precise nature in the dataset. We use control regions as surrogates that carry information regarding the bias, employ the classifier model to extract features, and suppress biased intermediate features with our custom, modular DecorreLayer. We evaluate our method on a dataset of 952 lung computed tomography scans by introducing simulated biases w.r.t. reconstruction kernel and noise level and propose including an adversarial test set in evaluations of bias reduction techniques. In a moderately sized model architecture, applying the proposed method to learn from data exhibiting a strong bias, it near-perfectly recovers the classification performance observed when training with corresponding unbiased data.
Building Brains: Subvolume Recombination for Data Augmentation in Large Vessel Occlusion Detection
May 16, 2022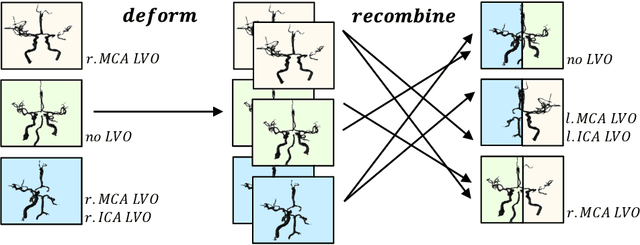
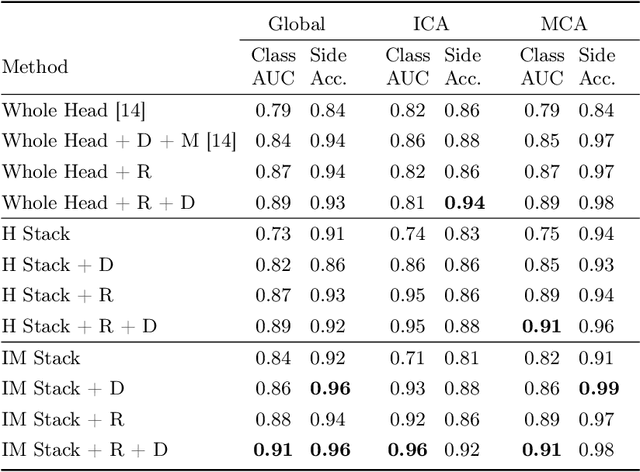
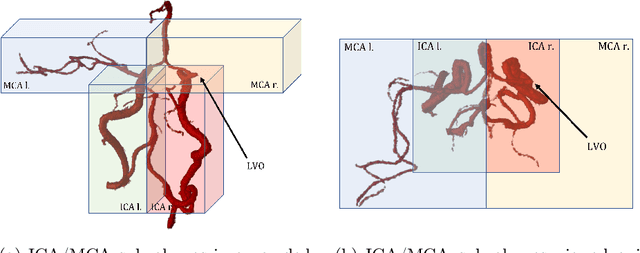
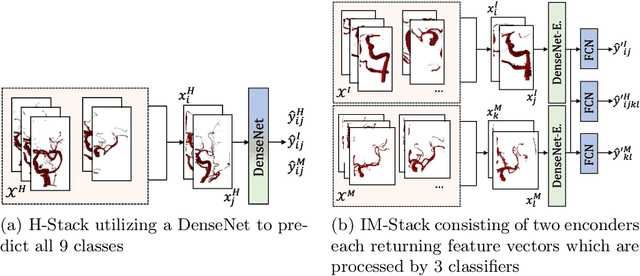
Abstract:Ischemic strokes are often caused by large vessel occlusions (LVOs), which can be visualized and diagnosed with Computed Tomography Angiography scans. As time is brain, a fast, accurate and automated diagnosis of these scans is desirable. Human readers compare the left and right hemispheres in their assessment of strokes. A large training data set is required for a standard deep learning-based model to learn this strategy from data. As labeled medical data in this field is rare, other approaches need to be developed. To both include the prior knowledge of side comparison and increase the amount of training data, we propose an augmentation method that generates artificial training samples by recombining vessel tree segmentations of the hemispheres or hemisphere subregions from different patients. The subregions cover vessels commonly affected by LVOs, namely the internal carotid artery (ICA) and middle cerebral artery (MCA). In line with the augmentation scheme, we use a 3D-DenseNet fed with task-specific input, fostering a side-by-side comparison between the hemispheres. Furthermore, we propose an extension of that architecture to process the individual hemisphere subregions. All configurations predict the presence of an LVO, its side, and the affected subregion. We show the effect of recombination as an augmentation strategy in a 5-fold cross validated ablation study. We enhanced the AUC for patient-wise classification regarding the presence of an LVO of all investigated architectures. For one variant, the proposed method improved the AUC from 0.73 without augmentation to 0.89. The best configuration detects LVOs with an AUC of 0.91, LVOs in the ICA with an AUC of 0.96, and in the MCA with 0.91 while accurately predicting the affected side.
CAD-RADS Scoring using Deep Learning and Task-Specific Centerline Labeling
Feb 08, 2022



Abstract:With coronary artery disease (CAD) persisting to be one of the leading causes of death worldwide, interest in supporting physicians with algorithms to speed up and improve diagnosis is high. In clinical practice, the severeness of CAD is often assessed with a coronary CT angiography (CCTA) scan and manually graded with the CAD-Reporting and Data System (CAD-RADS) score. The clinical questions this score assesses are whether patients have CAD or not (rule-out) and whether they have severe CAD or not (hold-out). In this work, we reach new state-of-the-art performance for automatic CAD-RADS scoring. We propose using severity-based label encoding, test time augmentation (TTA) and model ensembling for a task-specific deep learning architecture. Furthermore, we introduce a novel task- and model-specific, heuristic coronary segment labeling, which subdivides coronary trees into consistent parts across patients. It is fast, robust, and easy to implement. We were able to raise the previously reported area under the receiver operating characteristic curve (AUC) from 0.914 to 0.942 in the rule-out and from 0.921 to 0.950 in the hold-out task respectively.
Segmentation of the Carotid Lumen and Vessel Wall using Deep Learning and Location Priors
Jan 17, 2022
Abstract:In this report we want to present our method and results for the Carotid Artery Vessel Wall Segmentation Challenge. We propose an image-based pipeline utilizing the U-Net architecture and location priors to solve the segmentation problem at hand.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge