Maximilian Rohleder
Pattern Recognition Lab, FAU Erlangen-Nürnberg, Germany, Siemens Healthcare GmbH, Forchheim, Germany
A Realistic Collimated X-Ray Image Simulation Pipeline
Nov 15, 2024Abstract:Collimator detection remains a challenging task in X-ray systems with unreliable or non-available information about the detectors position relative to the source. This paper presents a physically motivated image processing pipeline for simulating the characteristics of collimator shadows in X-ray images. By generating randomized labels for collimator shapes and locations, incorporating scattered radiation simulation, and including Poisson noise, the pipeline enables the expansion of limited datasets for training deep neural networks. We validate the proposed pipeline by a qualitative and quantitative comparison against real collimator shadows. Furthermore, it is demonstrated that utilizing simulated data within our deep learning framework not only serves as a suitable substitute for actual collimators but also enhances the generalization performance when applied to real-world data.
Physics-Informed Learning for Time-Resolved Angiographic Contrast Agent Concentration Reconstruction
Mar 04, 2024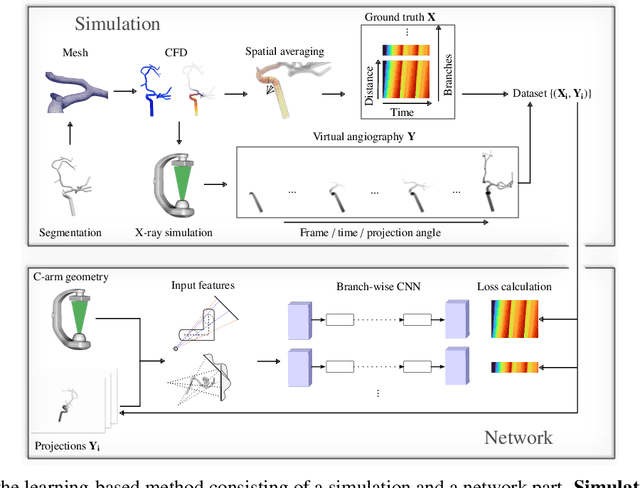
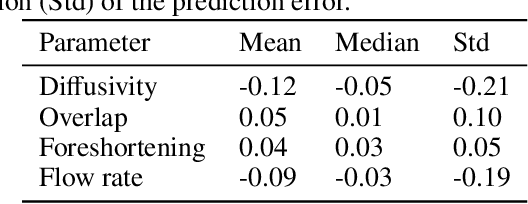
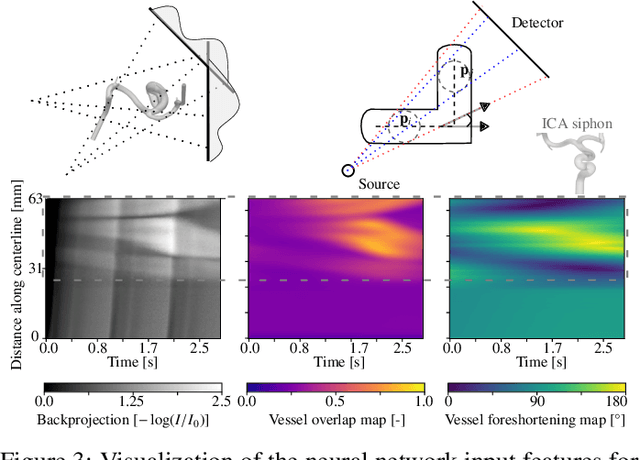
Abstract:Three-dimensional Digital Subtraction Angiography (3D-DSA) is a well-established X-ray-based technique for visualizing vascular anatomy. Recently, four-dimensional DSA (4D-DSA) reconstruction algorithms have been developed to enable the visualization of volumetric contrast flow dynamics through time-series of volumes. . This reconstruction problem is ill-posed mainly due to vessel overlap in the projection direction and geometric vessel foreshortening, which leads to information loss in the recorded projection images. However, knowledge about the underlying fluid dynamics can be leveraged to constrain the solution space. In our work, we implicitly include this information in a neural network-based model that is trained on a dataset of image-based blood flow simulations. The model predicts the spatially averaged contrast agent concentration for each centerline point of the vasculature over time, lowering the overall computational demand. The trained network enables the reconstruction of relative contrast agent concentrations with a mean absolute error of 0.02 $\pm$ 0.02 and a mean absolute percentage error of 5.31 % $\pm$ 9.25 %. Moreover, the network is robust to varying degrees of vessel overlap and vessel foreshortening. Our approach demonstrates the potential of the integration of machine learning and blood flow simulations in time-resolved angiographic flow reconstruction.
Transient Hemodynamics Prediction Using an Efficient Octree-Based Deep Learning Model
Feb 13, 2023
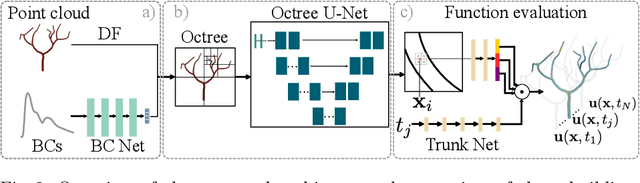
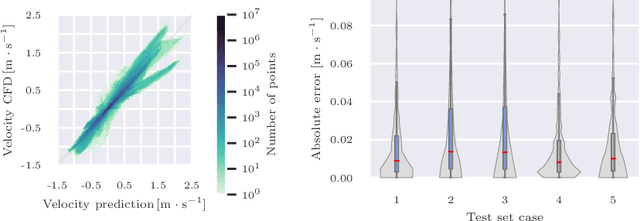
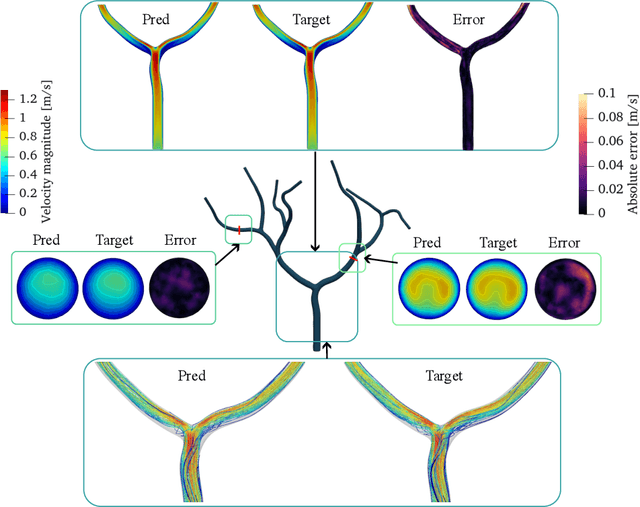
Abstract:Patient-specific hemodynamics assessment could support diagnosis and treatment of neurovascular diseases. Currently, conventional medical imaging modalities are not able to accurately acquire high-resolution hemodynamic information that would be required to assess complex neurovascular pathologies. Therefore, computational fluid dynamics (CFD) simulations can be applied to tomographic reconstructions to obtain clinically relevant information. However, three-dimensional (3D) CFD simulations require enormous computational resources and simulation-related expert knowledge that are usually not available in clinical environments. Recently, deep-learning-based methods have been proposed as CFD surrogates to improve computational efficiency. Nevertheless, the prediction of high-resolution transient CFD simulations for complex vascular geometries poses a challenge to conventional deep learning models. In this work, we present an architecture that is tailored to predict high-resolution (spatial and temporal) velocity fields for complex synthetic vascular geometries. For this, an octree-based spatial discretization is combined with an implicit neural function representation to efficiently handle the prediction of the 3D velocity field for each time step. The presented method is evaluated for the task of cerebral hemodynamics prediction before and during the injection of contrast agent in the internal carotid artery (ICA). Compared to CFD simulations, the velocity field can be estimated with a mean absolute error of 0.024 m/s, whereas the run time reduces from several hours on a high-performance cluster to a few seconds on a consumer graphical processing unit.
Gradient-Based Geometry Learning for Fan-Beam CT Reconstruction
Dec 05, 2022Abstract:Incorporating computed tomography (CT) reconstruction operators into differentiable pipelines has proven beneficial in many applications. Such approaches usually focus on the projection data and keep the acquisition geometry fixed. However, precise knowledge of the acquisition geometry is essential for high quality reconstruction results. In this paper, the differentiable formulation of fan-beam CT reconstruction is extended to the acquisition geometry. This allows to propagate gradient information from a loss function on the reconstructed image into the geometry parameters. As a proof-of-concept experiment, this idea is applied to rigid motion compensation. The cost function is parameterized by a trained neural network which regresses an image quality metric from the motion affected reconstruction alone. Using the proposed method, we are the first to optimize such an autofocus-inspired algorithm based on analytical gradients. The algorithm achieves a reduction in MSE by 35.5 % and an improvement in SSIM by 12.6 % over the motion affected reconstruction. Next to motion compensation, we see further use cases of our differentiable method for scanner calibration or hybrid techniques employing deep models.
On the Benefit of Dual-domain Denoising in a Self-supervised Low-dose CT Setting
Nov 03, 2022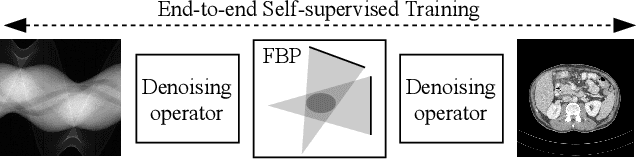
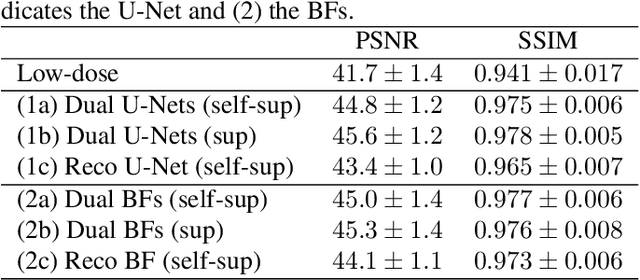
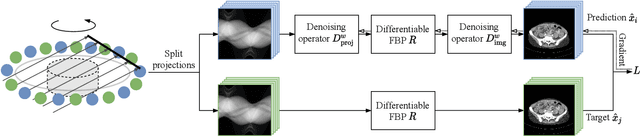
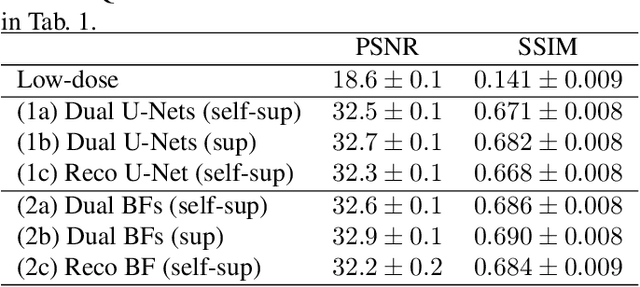
Abstract:Computed tomography (CT) is routinely used for three-dimensional non-invasive imaging. Numerous data-driven image denoising algorithms were proposed to restore image quality in low-dose acquisitions. However, considerably less research investigates methods already intervening in the raw detector data due to limited access to suitable projection data or correct reconstruction algorithms. In this work, we present an end-to-end trainable CT reconstruction pipeline that contains denoising operators in both the projection and the image domain and that are optimized simultaneously without requiring ground-truth high-dose CT data. Our experiments demonstrate that including an additional projection denoising operator improved the overall denoising performance by 82.4-94.1%/12.5-41.7% (PSNR/SSIM) on abdomen CT and 1.5-2.9%/0.4-0.5% (PSNR/SSIM) on XRM data relative to the low-dose baseline. We make our entire helical CT reconstruction framework publicly available that contains a raw projection rebinning step to render helical projection data suitable for differentiable fan-beam reconstruction operators and end-to-end learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge