Ludwig Ritschl
From Lines to Shapes: Geometric-Constrained Segmentation of X-Ray Collimators via Hough Transform
Sep 04, 2025Abstract:Collimation in X-ray imaging restricts exposure to the region-of-interest (ROI) and minimizes the radiation dose applied to the patient. The detection of collimator shadows is an essential image-based preprocessing step in digital radiography posing a challenge when edges get obscured by scattered X-ray radiation. Regardless, the prior knowledge that collimation forms polygonal-shaped shadows is evident. For this reason, we introduce a deep learning-based segmentation that is inherently constrained to its geometry. We achieve this by incorporating a differentiable Hough transform-based network to detect the collimation borders and enhance its capability to extract the information about the ROI center. During inference, we combine the information of both tasks to enable the generation of refined, line-constrained segmentation masks. We demonstrate robust reconstruction of collimated regions achieving median Hausdorff distances of 4.3-5.0mm on diverse test sets of real Xray images. While this application involves at most four shadow borders, our method is not fundamentally limited by a specific number of edges.
A Realistic Collimated X-Ray Image Simulation Pipeline
Nov 15, 2024Abstract:Collimator detection remains a challenging task in X-ray systems with unreliable or non-available information about the detectors position relative to the source. This paper presents a physically motivated image processing pipeline for simulating the characteristics of collimator shadows in X-ray images. By generating randomized labels for collimator shapes and locations, incorporating scattered radiation simulation, and including Poisson noise, the pipeline enables the expansion of limited datasets for training deep neural networks. We validate the proposed pipeline by a qualitative and quantitative comparison against real collimator shadows. Furthermore, it is demonstrated that utilizing simulated data within our deep learning framework not only serves as a suitable substitute for actual collimators but also enhances the generalization performance when applied to real-world data.
An Interpretable X-ray Style Transfer via Trainable Local Laplacian Filter
Nov 11, 2024



Abstract:Radiologists have preferred visual impressions or 'styles' of X-ray images that are manually adjusted to their needs to support their diagnostic performance. In this work, we propose an automatic and interpretable X-ray style transfer by introducing a trainable version of the Local Laplacian Filter (LLF). From the shape of the LLF's optimized remap function, the characteristics of the style transfer can be inferred and reliability of the algorithm can be ensured. Moreover, we enable the LLF to capture complex X-ray style features by replacing the remap function with a Multi-Layer Perceptron (MLP) and adding a trainable normalization layer. We demonstrate the effectiveness of the proposed method by transforming unprocessed mammographic X-ray images into images that match the style of target mammograms and achieve a Structural Similarity Index (SSIM) of 0.94 compared to 0.82 of the baseline LLF style transfer method from Aubry et al.
StyleX: A Trainable Metric for X-ray Style Distances
May 23, 2024Abstract:The progression of X-ray technology introduces diverse image styles that need to be adapted to the preferences of radiologists. To support this task, we introduce a novel deep learning-based metric that quantifies style differences of non-matching image pairs. At the heart of our metric is an encoder capable of generating X-ray image style representations. This encoder is trained without any explicit knowledge of style distances by exploiting Simple Siamese learning. During inference, the style representations produced by the encoder are used to calculate a distance metric for non-matching image pairs. Our experiments investigate the proposed concept for a disclosed reproducible and a proprietary image processing pipeline along two dimensions: First, we use a t-distributed stochastic neighbor embedding (t-SNE) analysis to illustrate that the encoder outputs provide meaningful and discriminative style representations. Second, the proposed metric calculated from the encoder outputs is shown to quantify style distances for non-matching pairs in good alignment with the human perception. These results confirm that our proposed method is a promising technique to quantify style differences, which can be used for guided style selection as well as automatic optimization of image pipeline parameters.
Metal-conscious Embedding for CBCT Projection Inpainting
Nov 29, 2022Abstract:The existence of metallic implants in projection images for cone-beam computed tomography (CBCT) introduces undesired artifacts which degrade the quality of reconstructed images. In order to reduce metal artifacts, projection inpainting is an essential step in many metal artifact reduction algorithms. In this work, a hybrid network combining the shift window (Swin) vision transformer (ViT) and a convolutional neural network is proposed as a baseline network for the inpainting task. To incorporate metal information for the Swin ViT-based encoder, metal-conscious self-embedding and neighborhood-embedding methods are investigated. Both methods have improved the performance of the baseline network. Furthermore, by choosing appropriate window size, the model with neighborhood-embedding could achieve the lowest mean absolute error of 0.079 in metal regions and the highest peak signal-to-noise ratio of 42.346 in CBCT projections. At the end, the efficiency of metal-conscious embedding on both simulated and real cadaver CBCT data has been demonstrated, where the inpainting capability of the baseline network has been enhanced.
Simulation-Driven Training of Vision Transformers Enabling Metal Segmentation in X-Ray Images
Mar 17, 2022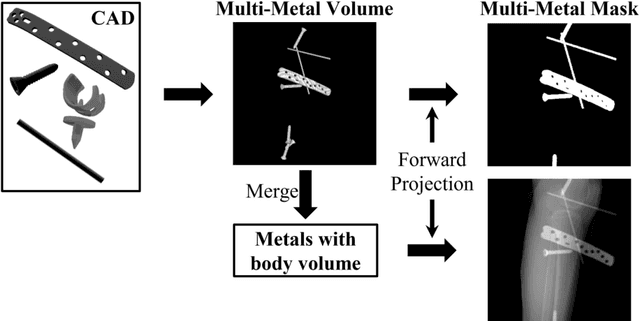
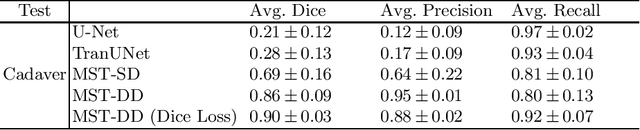
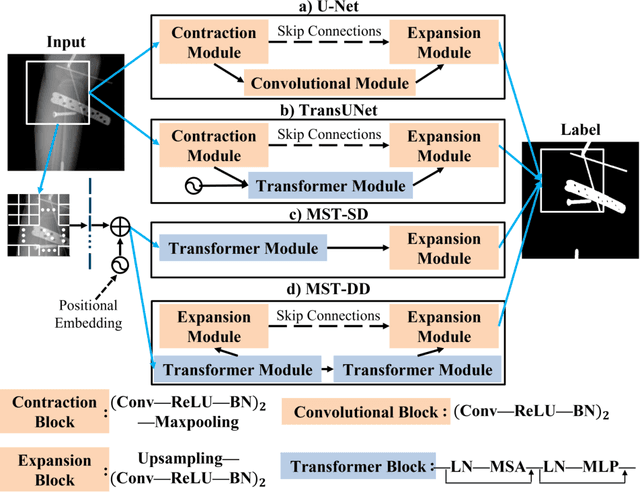
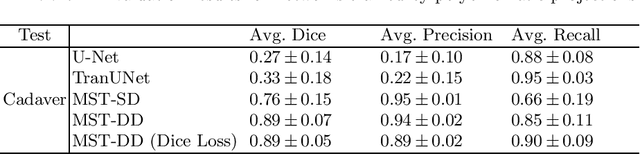
Abstract:In several image acquisition and processing steps of X-ray radiography, knowledge of the existence of metal implants and their exact position is highly beneficial (e.g. dose regulation, image contrast adjustment). Another application which would benefit from an accurate metal segmentation is cone beam computed tomography (CBCT) which is based on 2D X-ray projections. Due to the high attenuation of metals, severe artifacts occur in the 3D X-ray acquisitions. The metal segmentation in CBCT projections usually serves as a prerequisite for metal artifact avoidance and reduction algorithms. Since the generation of high quality clinical training is a constant challenge, this study proposes to generate simulated X-ray images based on CT data sets combined with self-designed computer aided design (CAD) implants and make use of convolutional neural network (CNN) and vision transformer (ViT) for metal segmentation. Model test is performed on accurately labeled X-ray test datasets obtained from specimen scans. The CNN encoder-based network like U-Net has limited performance on cadaver test data with an average dice score below 0.30, while the metal segmentation transformer with dual decoder (MST-DD) shows high robustness and generalization on the segmentation task, with an average dice score of 0.90. Our study indicates that the CAD model-based data generation has high flexibility and could be a way to overcome the problem of shortage in clinical data sampling and labelling. Furthermore, the MST-DD approach generates a more reliable neural network in case of training on simulated data.
Deep Learning-based Denoising of Mammographic Images using Physics-driven Data Augmentation
Dec 11, 2019


Abstract:Mammography is using low-energy X-rays to screen the human breast and is utilized by radiologists to detect breast cancer. Typically radiologists require a mammogram with impeccable image quality for an accurate diagnosis. In this study, we propose a deep learning method based on Convolutional Neural Networks (CNNs) for mammogram denoising to improve the image quality. We first enhance the noise level and employ Anscombe Transformation (AT) to transform Poisson noise to white Gaussian noise. With this data augmentation, a deep residual network is trained to learn the noise map of the noisy images. We show, that the proposed method can remove not only simulated but also real noise. Furthermore, we also compare our results with state-of-the-art denoising methods, such as BM3D and DNCNN. In an early investigation, we achieved qualitatively better mammogram denoising results.
Learning to recognize Abnormalities in Chest X-Rays with Location-Aware Dense Networks
Mar 12, 2018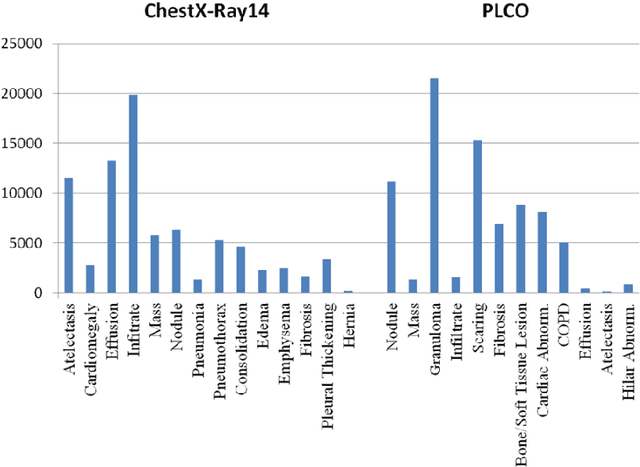
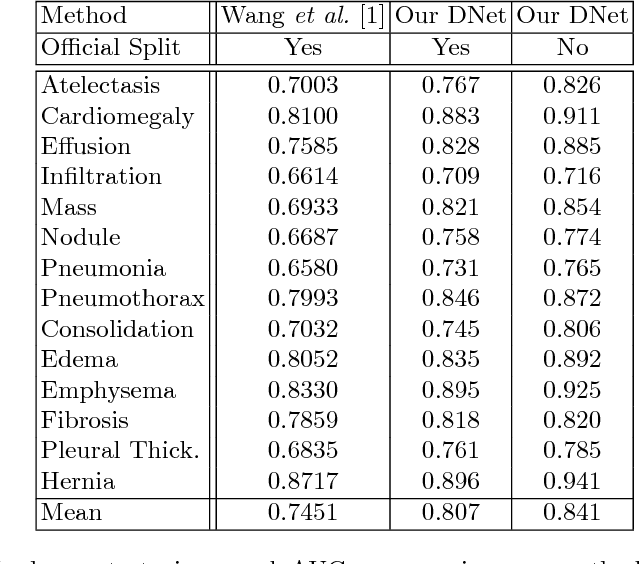
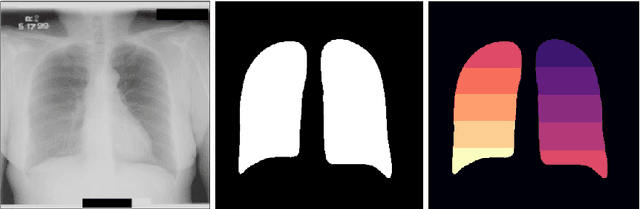

Abstract:Chest X-ray is the most common medical imaging exam used to assess multiple pathologies. Automated algorithms and tools have the potential to support the reading workflow, improve efficiency, and reduce reading errors. With the availability of large scale data sets, several methods have been proposed to classify pathologies on chest X-ray images. However, most methods report performance based on random image based splitting, ignoring the high probability of the same patient appearing in both training and test set. In addition, most methods fail to explicitly incorporate the spatial information of abnormalities or utilize the high resolution images. We propose a novel approach based on location aware Dense Networks (DNetLoc), whereby we incorporate both high-resolution image data and spatial information for abnormality classification. We evaluate our method on the largest data set reported in the community, containing a total of 86,876 patients and 297,541 chest X-ray images. We achieve (i) the best average AUC score for published training and test splits on the single benchmarking data set (ChestX-Ray14), and (ii) improved AUC scores when the pathology location information is explicitly used. To foster future research we demonstrate the limitations of the current benchmarking setup and provide new reference patient-wise splits for the used data sets. This could support consistent and meaningful benchmarking of future methods on the largest publicly available data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge