Sebastian Guendel
Extracting and Leveraging Nodule Features with Lung Inpainting for Local Feature Augmentation
Aug 05, 2020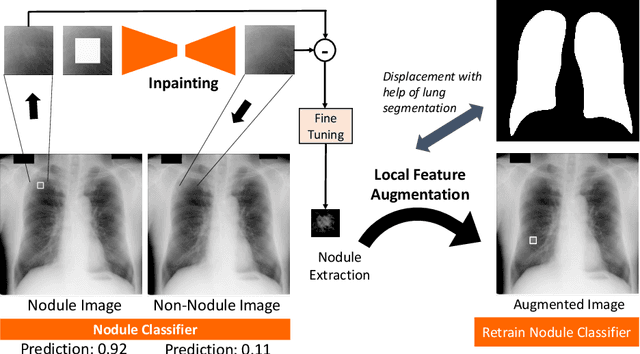
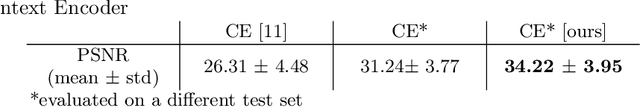
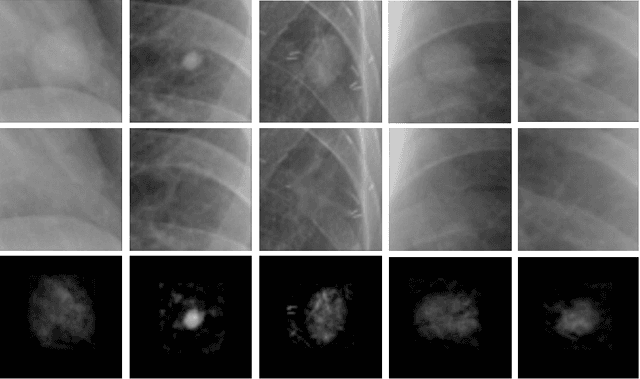
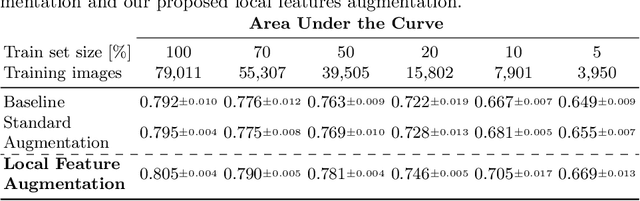
Abstract:Chest X-ray (CXR) is the most common examination for fast detection of pulmonary abnormalities. Recently, automated algorithms have been developed to classify multiple diseases and abnormalities in CXR scans. However, because of the limited availability of scans containing nodules and the subtle properties of nodules in CXRs, state-of-the-art methods do not perform well on nodule classification. To create additional data for the training process, standard augmentation techniques are applied. However, the variance introduced by these methods are limited as the images are typically modified globally. In this paper, we propose a method for local feature augmentation by extracting local nodule features using a generative inpainting network. The network is applied to generate realistic, healthy tissue and structures in patches containing nodules. The nodules are entirely removed in the inpainted representation. The extraction of the nodule features is processed by subtraction of the inpainted patch from the nodule patch. With arbitrary displacement of the extracted nodules in the lung area across different CXR scans and further local modifications during training, we significantly increase the nodule classification performance and outperform state-of-the-art augmentation methods.
Epoch-wise label attacks for robustness against label noise
Dec 12, 2019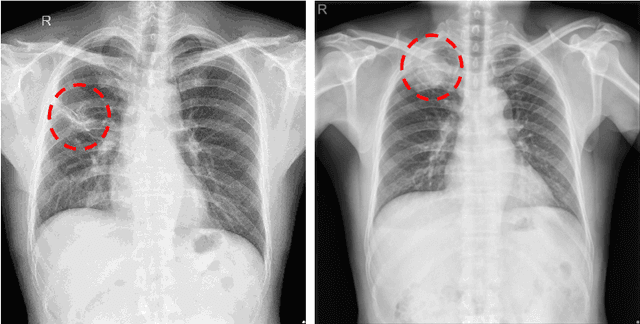

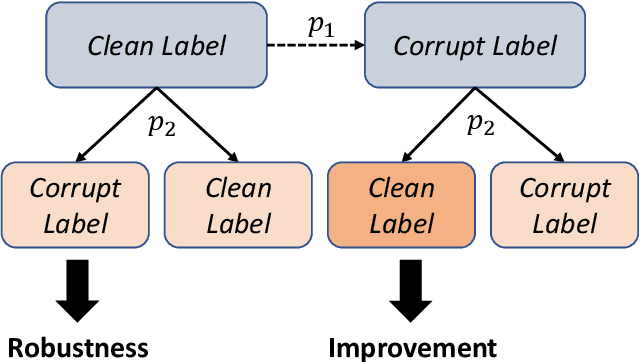

Abstract:The current accessibility to large medical datasets for training convolutional neural networks is tremendously high. The associated dataset labels are always considered to be the real "ground truth". However, the labeling procedures often seem to be inaccurate and many wrong labels are integrated. This may have fatal consequences on the performance of both training and evaluation. In this paper, we show the impact of label noise in the training set on a specific medical problem based on chest X-ray images. With a simple one-class problem, the classification of tuberculosis, we measure the performance on a clean evaluation set when training with label-corrupt data. We develop a method to compete with incorrectly labeled data during training by randomly attacking labels on individual epochs. The network tends to be robust when flipping correct labels for a single epoch and initiates a good step to the optimal minimum on the error surface when flipping noisy labels. On a baseline with an AUC (Area under Curve) score of 0.924, the performance drops to 0.809 when 30% of our training data is misclassified. With our approach the baseline performance could almost be maintained, the performance raised to 0.918.
Quantifying and Leveraging Classification Uncertainty for Chest Radiograph Assessment
Jun 18, 2019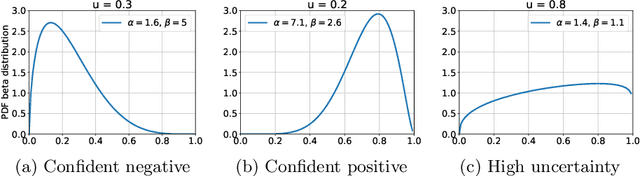
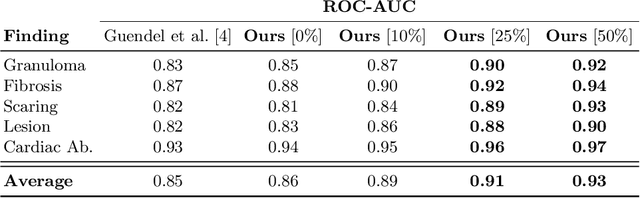
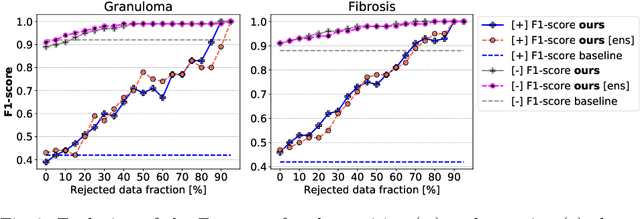
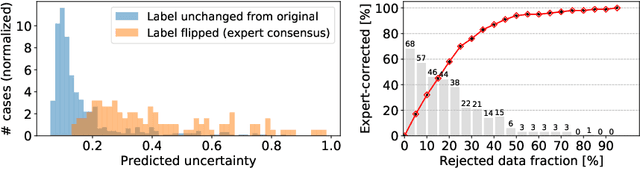
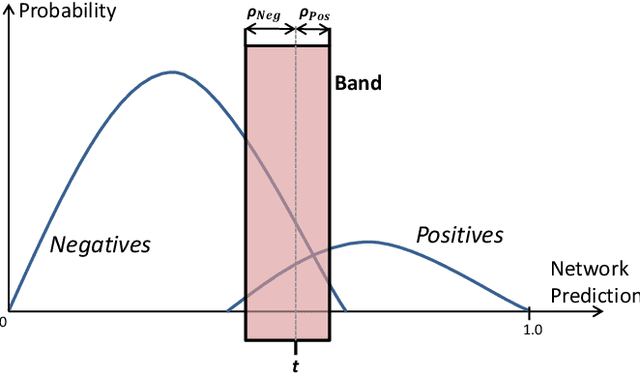
Abstract:The interpretation of chest radiographs is an essential task for the detection of thoracic diseases and abnormalities. However, it is a challenging problem with high inter-rater variability and inherent ambiguity due to inconclusive evidence in the data, limited data quality or subjective definitions of disease appearance. Current deep learning solutions for chest radiograph abnormality classification are typically limited to providing probabilistic predictions, relying on the capacity of learning models to adapt to the high degree of label noise and become robust to the enumerated causal factors. In practice, however, this leads to overconfident systems with poor generalization on unseen data. To account for this, we propose an automatic system that learns not only the probabilistic estimate on the presence of an abnormality, but also an explicit uncertainty measure which captures the confidence of the system in the predicted output. We argue that explicitly learning the classification uncertainty as an orthogonal measure to the predicted output, is essential to account for the inherent variability characteristic of this data. Experiments were conducted on two datasets of chest radiographs of over 85,000 patients. Sample rejection based on the predicted uncertainty can significantly improve the ROC-AUC, e.g., by 8% to 0.91 with an expected rejection rate of under 25%. Eliminating training samples using uncertainty-driven bootstrapping, enables a significant increase in robustness and accuracy. In addition, we present a multi-reader study showing that the predictive uncertainty is indicative of reader errors.
Multi-task Learning for Chest X-ray Abnormality Classification on Noisy Labels
May 15, 2019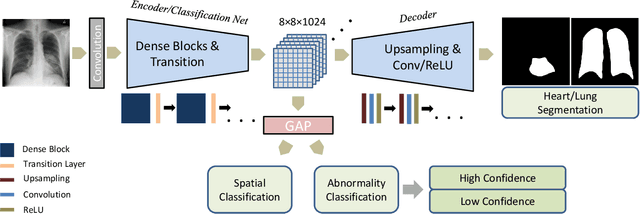
Abstract:Chest X-ray (CXR) is the most common X-ray examination performed in daily clinical practice for the diagnosis of various heart and lung abnormalities. The large amount of data to be read and reported, with 100+ studies per day for a single radiologist, poses a challenge in maintaining consistently high interpretation accuracy. In this work, we propose a method for the classification of different abnormalities based on CXR scans of the human body. The system is based on a novel multi-task deep learning architecture that in addition to the abnormality classification, supports the segmentation of the lungs and heart and classification of regions where the abnormality is located. We demonstrate that by training these tasks concurrently, one can increase the classification performance of the model. Experiments were performed on an extensive collection of 297,541 chest X-ray images from 86,876 patients, leading to a state-of-the-art performance level of 0.883 AUC on average for 12 different abnormalities. We also conducted a detailed performance analysis and compared the accuracy of our system with 3 board-certified radiologists. In this context, we highlight the high level of label noise inherent to this problem. On a reduced subset containing only cases with high confidence reference labels based on the consensus of the 3 radiologists, our system reached an average AUC of 0.945.
Learning to recognize Abnormalities in Chest X-Rays with Location-Aware Dense Networks
Mar 12, 2018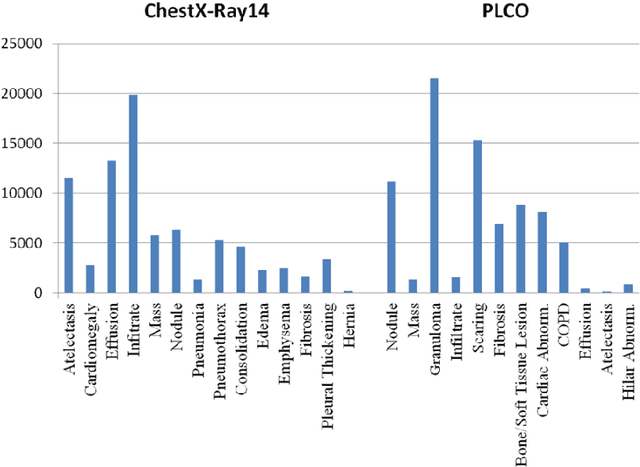
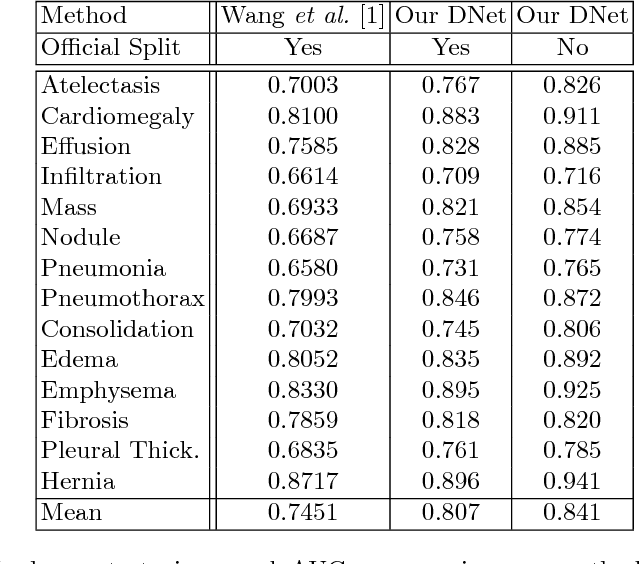
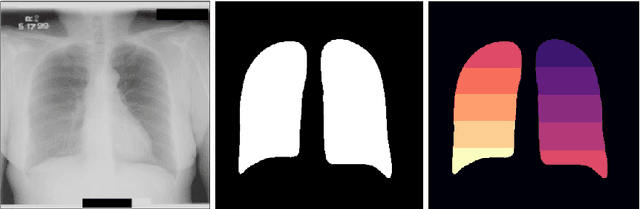

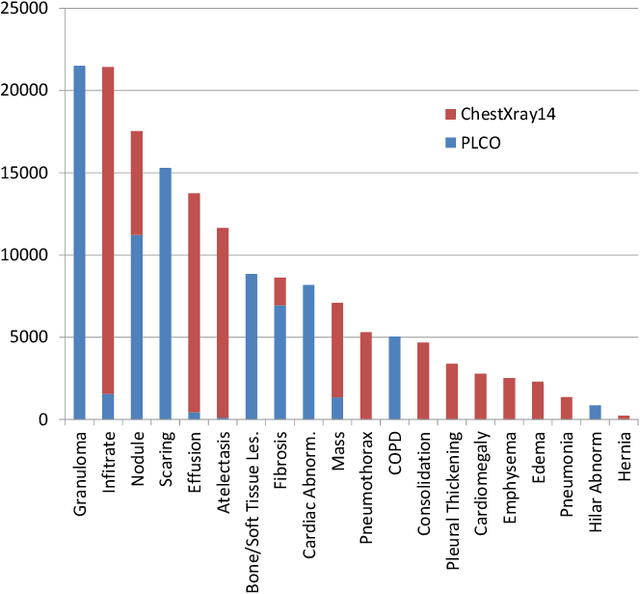
Abstract:Chest X-ray is the most common medical imaging exam used to assess multiple pathologies. Automated algorithms and tools have the potential to support the reading workflow, improve efficiency, and reduce reading errors. With the availability of large scale data sets, several methods have been proposed to classify pathologies on chest X-ray images. However, most methods report performance based on random image based splitting, ignoring the high probability of the same patient appearing in both training and test set. In addition, most methods fail to explicitly incorporate the spatial information of abnormalities or utilize the high resolution images. We propose a novel approach based on location aware Dense Networks (DNetLoc), whereby we incorporate both high-resolution image data and spatial information for abnormality classification. We evaluate our method on the largest data set reported in the community, containing a total of 86,876 patients and 297,541 chest X-ray images. We achieve (i) the best average AUC score for published training and test splits on the single benchmarking data set (ChestX-Ray14), and (ii) improved AUC scores when the pathology location information is explicitly used. To foster future research we demonstrate the limitations of the current benchmarking setup and provide new reference patient-wise splits for the used data sets. This could support consistent and meaningful benchmarking of future methods on the largest publicly available data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge