Alexander Mühlberg
Siemens Healthcare GmbH, Forchheim, Germany
Digital staining in optical microscopy using deep learning -- a review
Mar 14, 2023Abstract:Until recently, conventional biochemical staining had the undisputed status as well-established benchmark for most biomedical problems related to clinical diagnostics, fundamental research and biotechnology. Despite this role as gold-standard, staining protocols face several challenges, such as a need for extensive, manual processing of samples, substantial time delays, altered tissue homeostasis, limited choice of contrast agents for a given sample, 2D imaging instead of 3D tomography and many more. Label-free optical technologies, on the other hand, do not rely on exogenous and artificial markers, by exploiting intrinsic optical contrast mechanisms, where the specificity is typically less obvious to the human observer. Over the past few years, digital staining has emerged as a promising concept to use modern deep learning for the translation from optical contrast to established biochemical contrast of actual stainings. In this review article, we provide an in-depth analysis of the current state-of-the-art in this field, suggest methods of good practice, identify pitfalls and challenges and postulate promising advances towards potential future implementations and applications.
SEMPAI: a Self-Enhancing Multi-Photon Artificial Intelligence for prior-informed assessment of muscle function and pathology
Oct 28, 2022Abstract:Deep learning (DL) shows notable success in biomedical studies. However, most DL algorithms work as a black box, exclude biomedical experts, and need extensive data. We introduce the Self-Enhancing Multi-Photon Artificial Intelligence (SEMPAI), that integrates hypothesis-driven priors in a data-driven DL approach for research on multiphoton microscopy (MPM) of muscle fibers. SEMPAI utilizes meta-learning to optimize prior integration, data representation, and neural network architecture simultaneously. This allows hypothesis testing and provides interpretable feedback about the origin of biological information in MPM images. SEMPAI performs joint learning of several tasks to enable prediction for small datasets. The method is applied on an extensive multi-study dataset resulting in the largest joint analysis of pathologies and function for single muscle fibers. SEMPAI outperforms state-of-the-art biomarkers in six of seven predictive tasks, including those with scarce data. SEMPAI's DL models with integrated priors are superior to those without priors and to prior-only machine learning approaches.
DeepTechnome: Mitigating Unknown Bias in Deep Learning Based Assessment of CT Images
May 26, 2022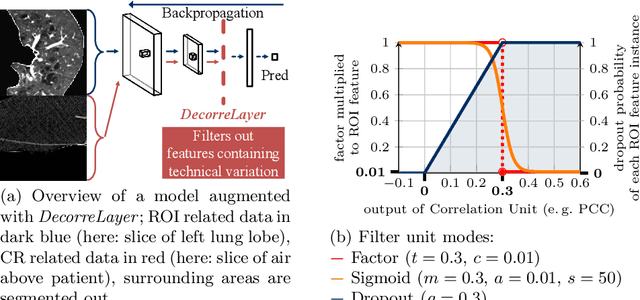
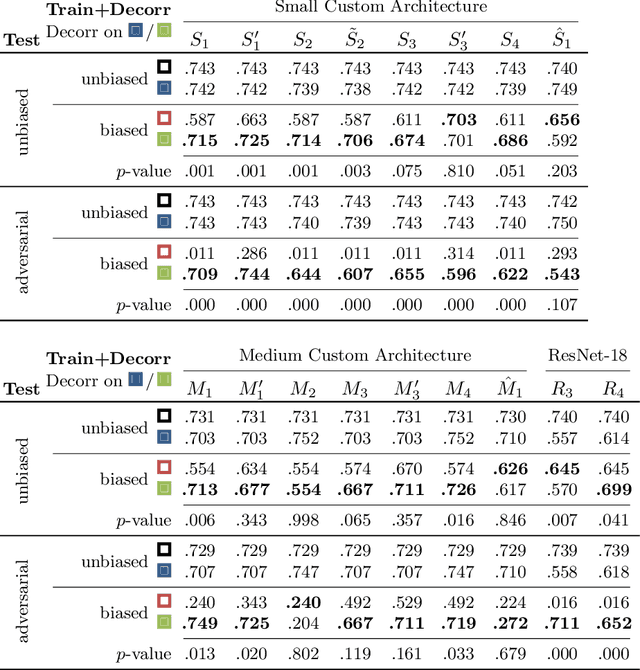

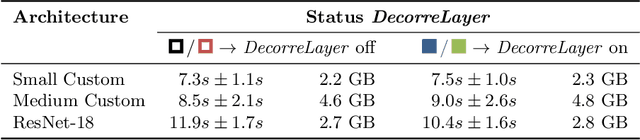
Abstract:Reliably detecting diseases using relevant biological information is crucial for real-world applicability of deep learning techniques in medical imaging. We debias deep learning models during training against unknown bias - without preprocessing/filtering the input beforehand or assuming specific knowledge about its distribution or precise nature in the dataset. We use control regions as surrogates that carry information regarding the bias, employ the classifier model to extract features, and suppress biased intermediate features with our custom, modular DecorreLayer. We evaluate our method on a dataset of 952 lung computed tomography scans by introducing simulated biases w.r.t. reconstruction kernel and noise level and propose including an adversarial test set in evaluations of bias reduction techniques. In a moderately sized model architecture, applying the proposed method to learn from data exhibiting a strong bias, it near-perfectly recovers the classification performance observed when training with corresponding unbiased data.
Explaining Clinical Decision Support Systems in Medical Imaging using Cycle-Consistent Activation Maximization
Oct 13, 2020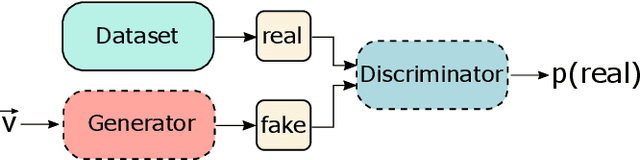
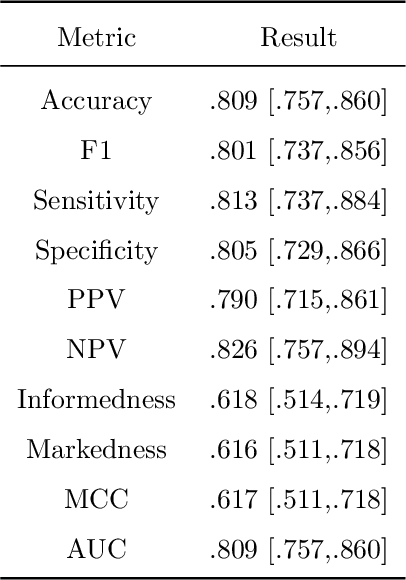
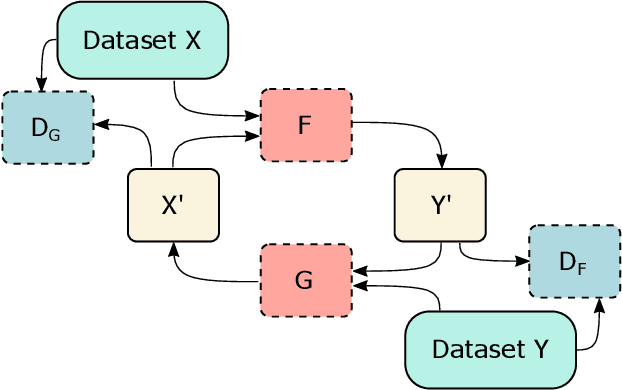
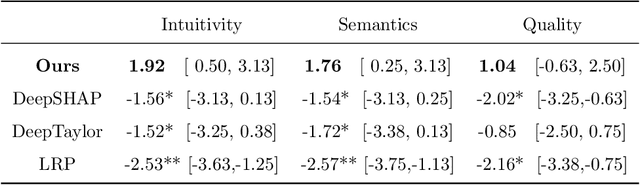
Abstract:Clinical decision support using deep neural networks has become a topic of steadily growing interest. While recent work has repeatedly demonstrated that deep learning offers major advantages for medical image classification over traditional methods, clinicians are often hesitant to adopt the technology because its underlying decision-making process is considered to be intransparent and difficult to comprehend. In recent years, this has been addressed by a variety of approaches that have successfully contributed to providing deeper insight. Most notably, additive feature attribution methods are able to propagate decisions back into the input space by creating a saliency map which allows the practitioner to "see what the network sees." However, the quality of the generated maps can become poor and the images noisy if only limited data is available - a typical scenario in clinical contexts. We propose a novel decision explanation scheme based on CycleGAN activation maximization which generates high-quality visualizations of classifier decisions even in smaller data sets. We conducted a user study in which these visualizations significantly outperformed existing methods on the LIDC dataset for lung lesion malignancy classification. With our approach we make a significant contribution to a better understanding of clinical decision support systems based on deep neural networks and thus aim to foster overall clinical acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge