Shiqi Xu
Enhancing the Efficiency and Accuracy of Underlying Asset Reviews in Structured Finance: The Application of Multi-agent Framework
May 07, 2024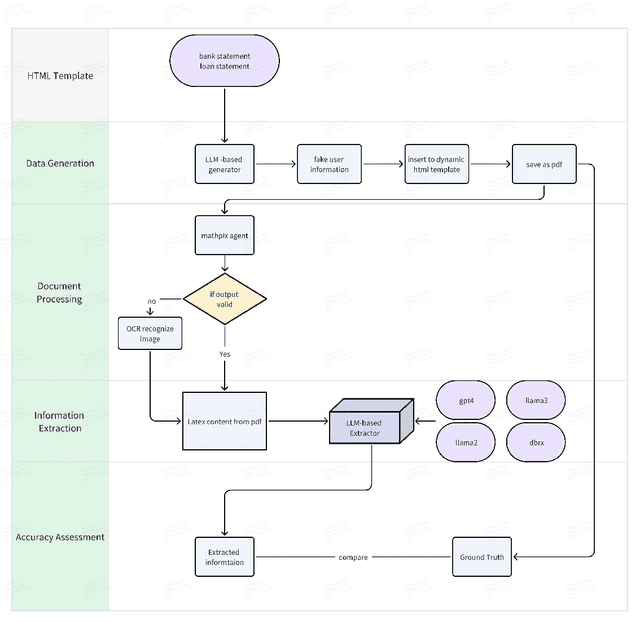
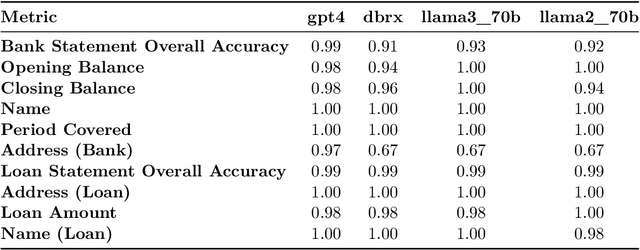
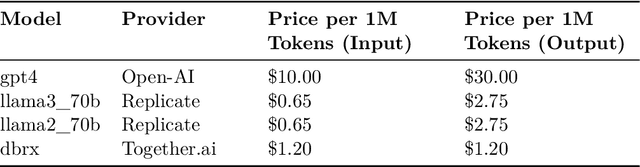
Abstract:Structured finance, which involves restructuring diverse assets into securities like MBS, ABS, and CDOs, enhances capital market efficiency but presents significant due diligence challenges. This study explores the integration of artificial intelligence (AI) with traditional asset review processes to improve efficiency and accuracy in structured finance. Using both open-sourced and close-sourced large language models (LLMs), we demonstrate that AI can automate the verification of information between loan applications and bank statements effectively. While close-sourced models such as GPT-4 show superior performance, open-sourced models like LLAMA3 offer a cost-effective alternative. Dual-agent systems further increase accuracy, though this comes with higher operational costs. This research highlights AI's potential to minimize manual errors and streamline due diligence, suggesting a broader application of AI in financial document analysis and risk management.
Inherent limitations of LLMs regarding spatial information
Dec 05, 2023



Abstract:Despite the significant advancements in natural language processing capabilities demonstrated by large language models such as ChatGPT, their proficiency in comprehending and processing spatial information, especially within the domains of 2D and 3D route planning, remains notably underdeveloped. This paper investigates the inherent limitations of ChatGPT and similar models in spatial reasoning and navigation-related tasks, an area critical for applications ranging from autonomous vehicle guidance to assistive technologies for the visually impaired. In this paper, we introduce a novel evaluation framework complemented by a baseline dataset, meticulously crafted for this study. This dataset is structured around three key tasks: plotting spatial points, planning routes in two-dimensional (2D) spaces, and devising pathways in three-dimensional (3D) environments. We specifically developed this dataset to assess the spatial reasoning abilities of ChatGPT. Our evaluation reveals key insights into the model's capabilities and limitations in spatial understanding.
Tensorial tomographic Fourier Ptychography with applications to muscle tissue imaging
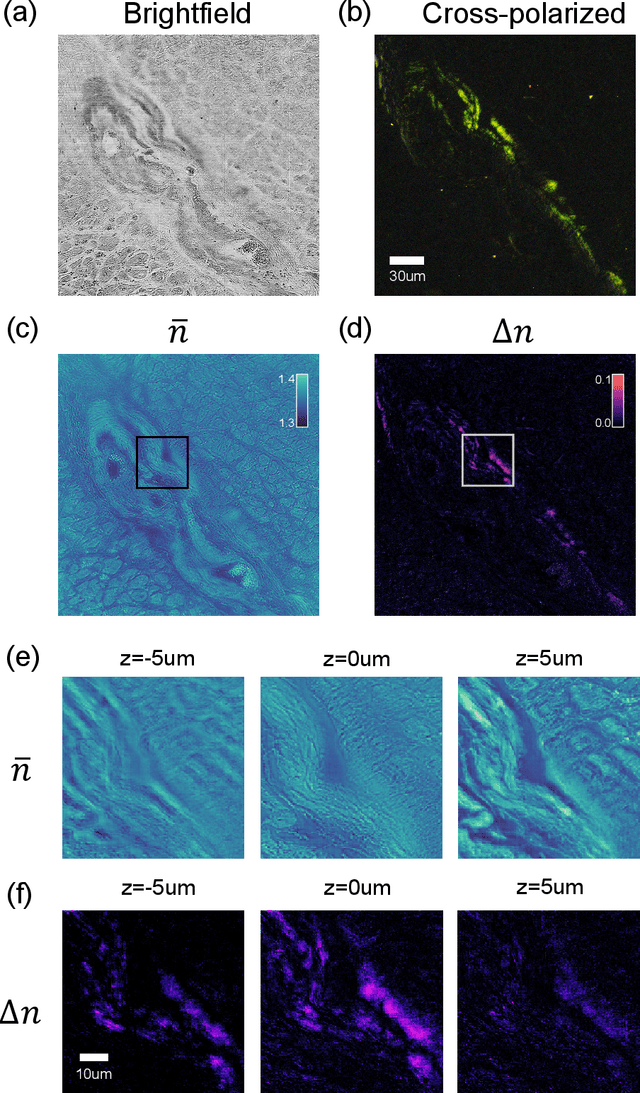
May 14, 2023Abstract:We report Tensorial tomographic Fourier Ptychography (ToFu), a new non-scanning label-free tomographic microscopy method for simultaneous imaging of quantitative phase and anisotropic specimen information in 3D. Built upon Fourier Ptychography, a quantitative phase imaging technique, ToFu additionally highlights the vectorial nature of light. The imaging setup consists of a standard microscope equipped with an LED matrix, a polarization generator, and a polarization-sensitive camera. Permittivity tensors of anisotropic samples are computationally recovered from polarized intensity measurements across three dimensions. We demonstrate ToFu's efficiency through volumetric reconstructions of refractive index, birefringence, and orientation for various validation samples, as well as tissue samples from muscle fibers and diseased heart tissue. Our reconstructions of muscle fibers resolve their 3D fine-filament structure and yield consistent morphological measurements compared to gold-standard second harmonic generation scanning confocal microscope images found in the literature. Additionally, we demonstrate reconstructions of a heart tissue sample that carries important polarization information for detecting cardiac amyloidosis.
Digital staining in optical microscopy using deep learning -- a review
Mar 14, 2023Abstract:Until recently, conventional biochemical staining had the undisputed status as well-established benchmark for most biomedical problems related to clinical diagnostics, fundamental research and biotechnology. Despite this role as gold-standard, staining protocols face several challenges, such as a need for extensive, manual processing of samples, substantial time delays, altered tissue homeostasis, limited choice of contrast agents for a given sample, 2D imaging instead of 3D tomography and many more. Label-free optical technologies, on the other hand, do not rely on exogenous and artificial markers, by exploiting intrinsic optical contrast mechanisms, where the specificity is typically less obvious to the human observer. Over the past few years, digital staining has emerged as a promising concept to use modern deep learning for the translation from optical contrast to established biochemical contrast of actual stainings. In this review article, we provide an in-depth analysis of the current state-of-the-art in this field, suggest methods of good practice, identify pitfalls and challenges and postulate promising advances towards potential future implementations and applications.
Multi-scale gigapixel microscopy using a multi-camera array microscope
Nov 30, 2022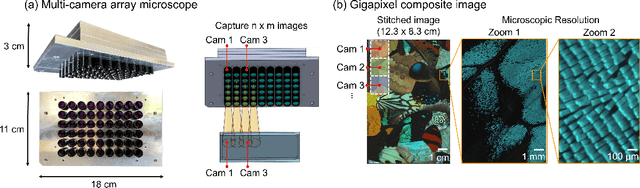
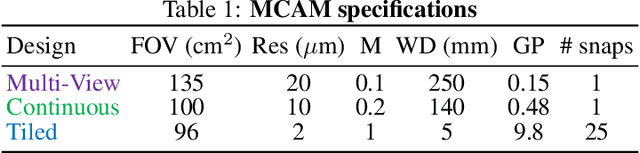
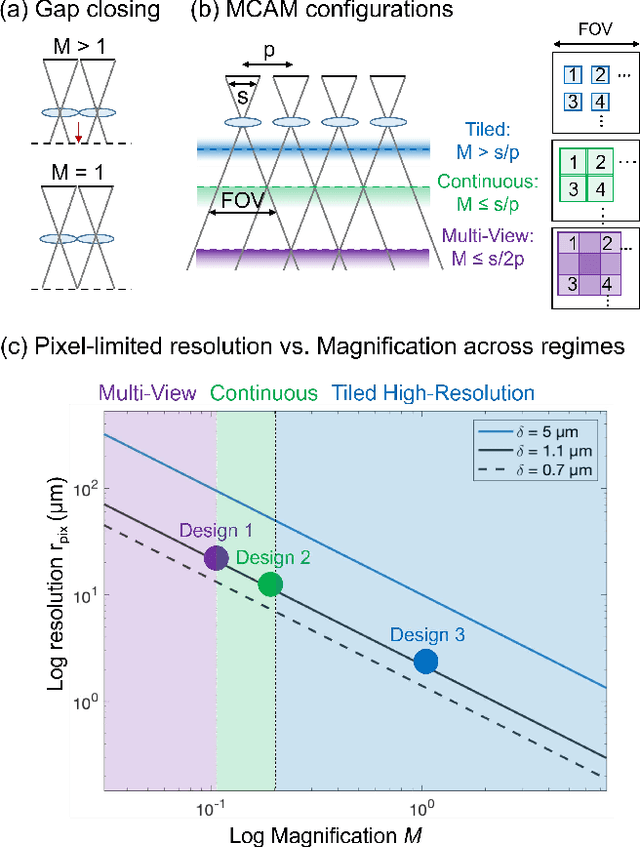
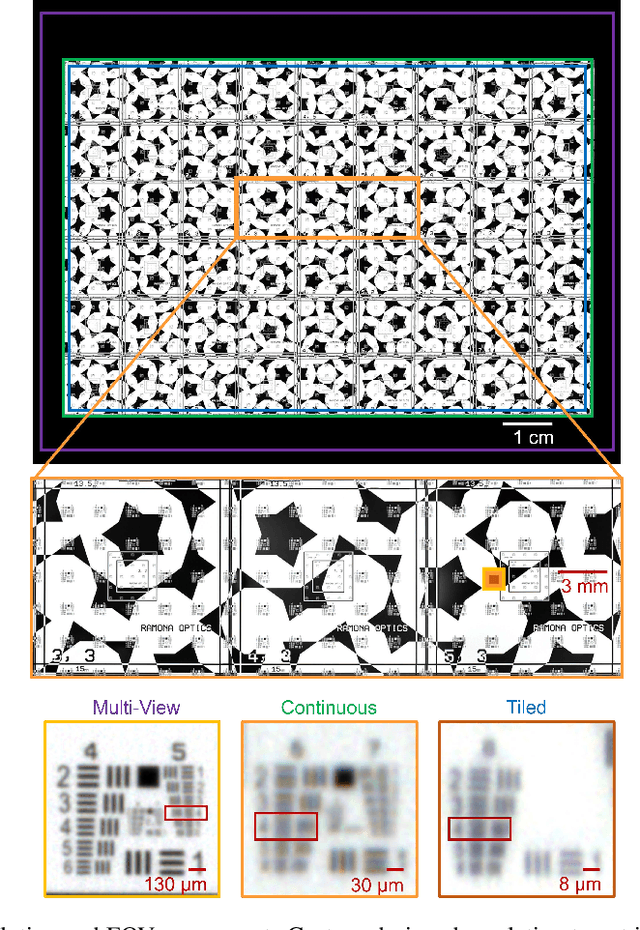
Abstract:This article experimentally examines different configurations of a novel multi-camera array microscope (MCAM) imaging technology. The MCAM is based upon a densely packed array of "micro-cameras" to jointly image across a large field-of-view at high resolution. Each micro-camera within the array images a unique area of a sample of interest, and then all acquired data with 54 micro-cameras are digitally combined into composite frames, whose total pixel counts significantly exceed the pixel counts of standard microscope systems. We present results from three unique MCAM configurations for different use cases. First, we demonstrate a configuration that simultaneously images and estimates the 3D object depth across a 100 x 135 mm^2 field-of-view (FOV) at approximately 20 um resolution, which results in 0.15 gigapixels (GP) per snapshot. Second, we demonstrate an MCAM configuration that records video across a continuous 83 x 123 mm^2 FOV with two-fold increased resolution (0.48 GP per frame). Finally, we report a third high-resolution configuration (2 um resolution) that can rapidly produce 9.8 GP composites of large histopathology specimens.
Tensorial tomographic differential phase-contrast microscopy
Apr 25, 2022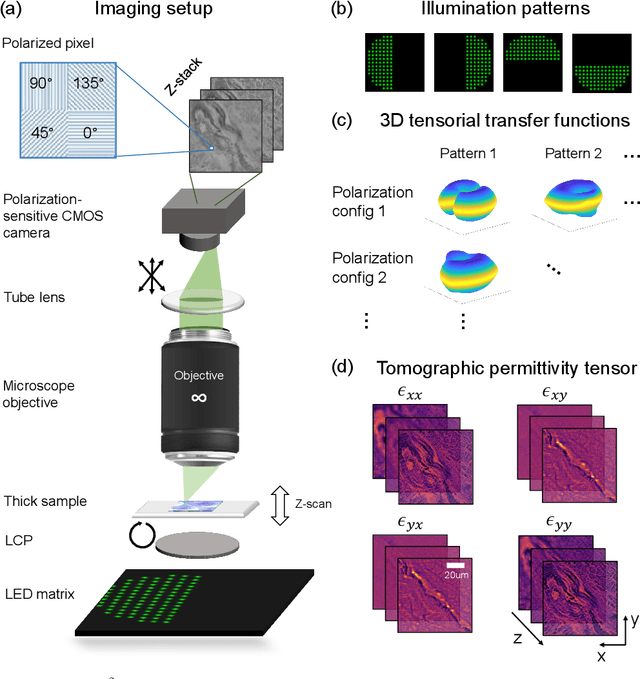
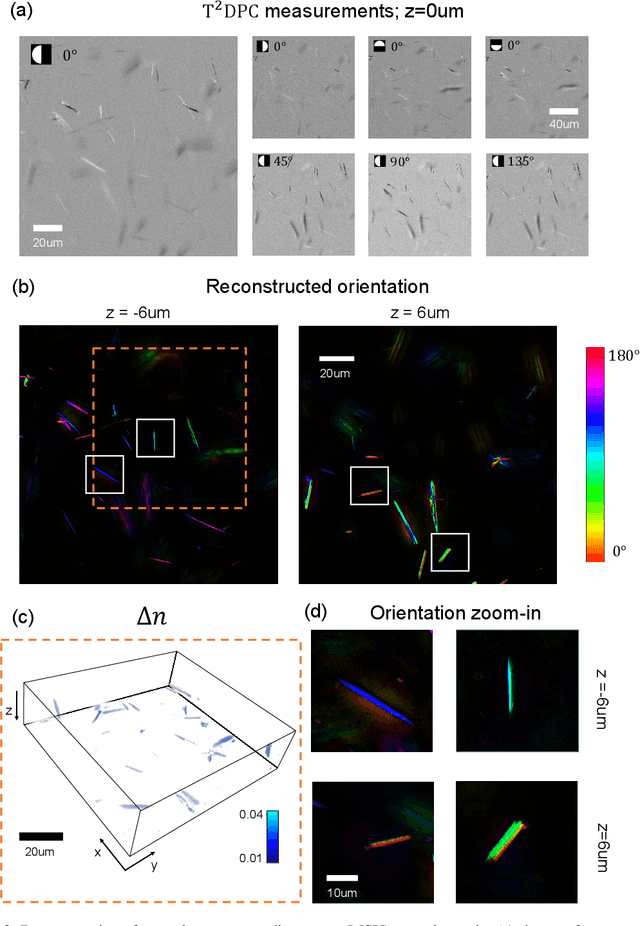
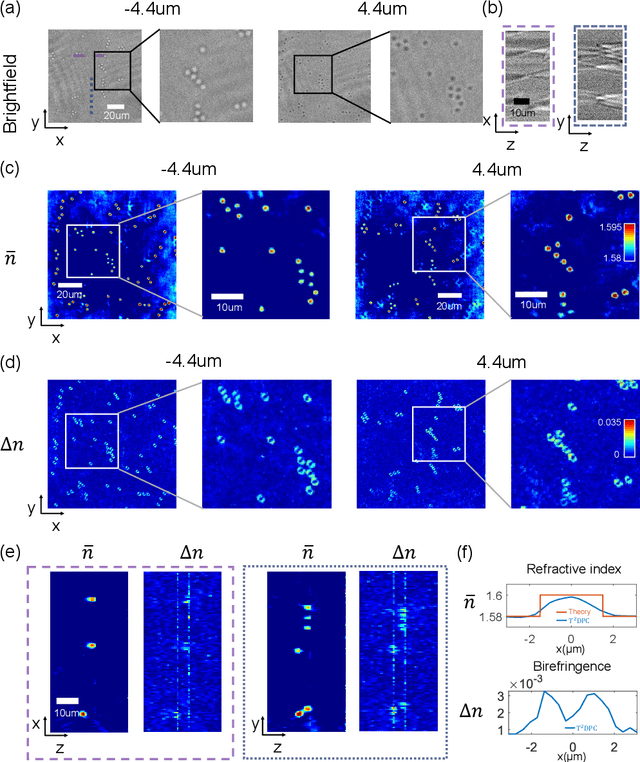
Abstract:We report Tensorial Tomographic Differential Phase-Contrast microscopy (T2DPC), a quantitative label-free tomographic imaging method for simultaneous measurement of phase and anisotropy. T2DPC extends differential phase-contrast microscopy, a quantitative phase imaging technique, to highlight the vectorial nature of light. The method solves for permittivity tensor of anisotropic samples from intensity measurements acquired with a standard microscope equipped with an LED matrix, a circular polarizer, and a polarization-sensitive camera. We demonstrate accurate volumetric reconstructions of refractive index, birefringence, and orientation for various validation samples, and show that the reconstructed polarization structures of a biological specimen are predictive of pathology.
Transient motion classification through turbid volumes via parallelized single-photon detection and deep contrastive embedding
Apr 04, 2022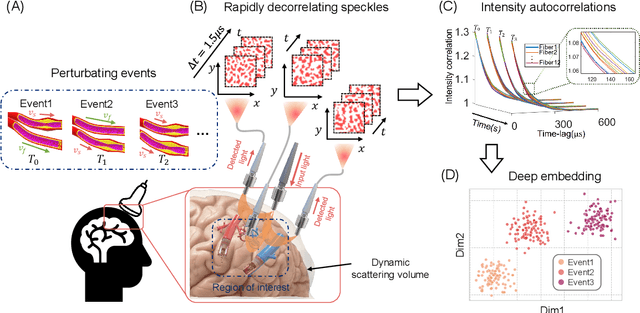
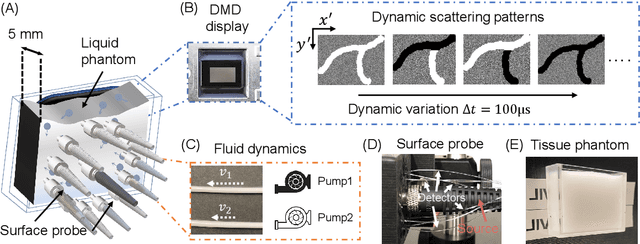
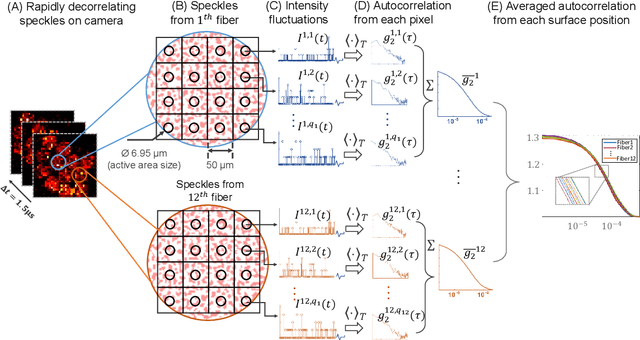
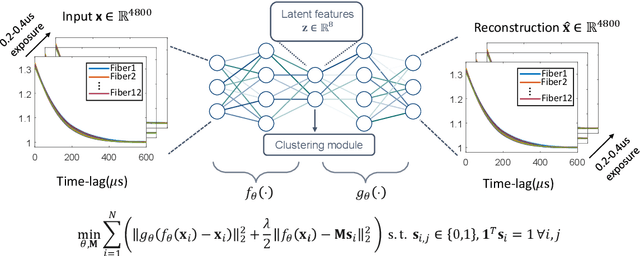
Abstract:Fast noninvasive probing of spatially varying decorrelating events, such as cerebral blood flow beneath the human skull, is an essential task in various scientific and clinical settings. One of the primary optical techniques used is diffuse correlation spectroscopy (DCS), whose classical implementation uses a single or few single-photon detectors, resulting in poor spatial localization accuracy and relatively low temporal resolution. Here, we propose a technique termed Classifying Rapid decorrelation Events via Parallelized single photon dEtection (CREPE)}, a new form of DCS that can probe and classify different decorrelating movements hidden underneath turbid volume with high sensitivity using parallelized speckle detection from a $32\times32$ pixel SPAD array. We evaluate our setup by classifying different spatiotemporal-decorrelating patterns hidden beneath a 5mm tissue-like phantom made with rapidly decorrelating dynamic scattering media. Twelve multi-mode fibers are used to collect scattered light from different positions on the surface of the tissue phantom. To validate our setup, we generate perturbed decorrelation patterns by both a digital micromirror device (DMD) modulated at multi-kilo-hertz rates, as well as a vessel phantom containing flowing fluid. Along with a deep contrastive learning algorithm that outperforms classic unsupervised learning methods, we demonstrate our approach can accurately detect and classify different transient decorrelation events (happening in 0.1-0.4s) underneath turbid scattering media, without any data labeling. This has the potential to be applied to noninvasively monitor deep tissue motion patterns, for example identifying normal or abnormal cerebral blood flow events, at multi-Hertz rates within a compact and static detection probe.
Quantitative Jones matrix imaging using vectorial Fourier ptychography
Oct 15, 2021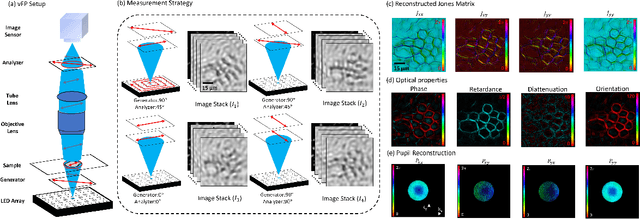
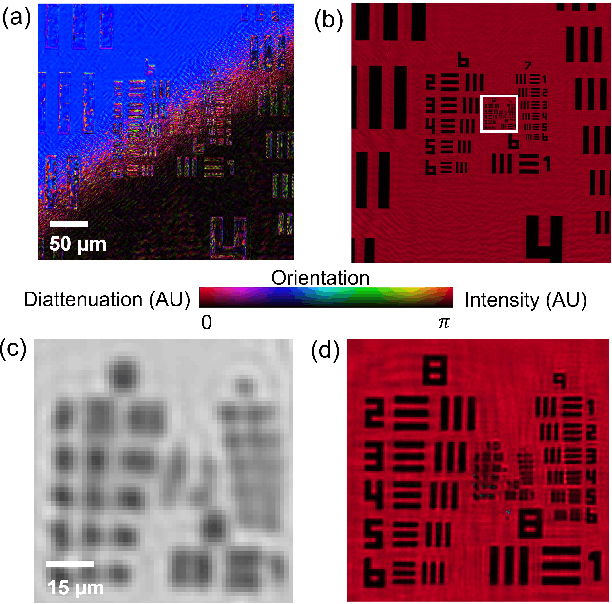
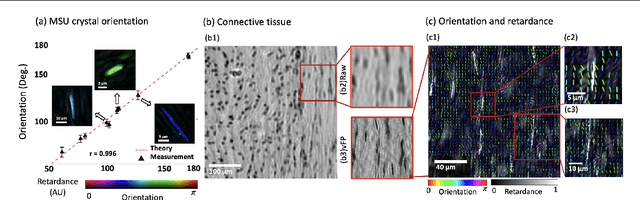
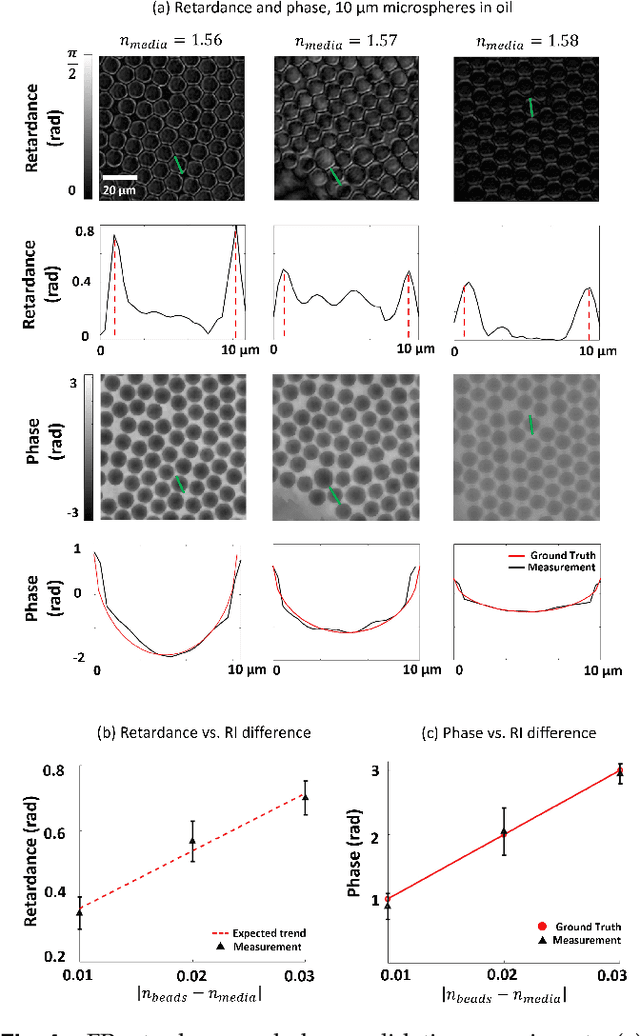
Abstract:This paper presents a microscopic imaging technique that uses variable-angle illumination to recover the complex polarimetric properties of a specimen at high resolution and over a large field-of-view. The approach extends Fourier ptychography, which is a synthetic aperture-based imaging approach to improve resolution with phaseless measurements, to additionally account for the vectorial nature of light. After images are acquired using a standard microscope outfitted with an LED illumination array and two polarizers, our vectorial Fourier Ptychography (vFP) algorithm solves for the complex 2x2 Jones matrix of the anisotropic specimen of interest at each resolved spatial location. We introduce a new sequential Gauss-Newton-based solver that additionally jointly estimates and removes polarization-dependent imaging system aberrations. We demonstrate effective vFP performance by generating large-area (29 mm$^2$), high-resolution (1.24 $\mu$m full-pitch) reconstructions of sample absorption, phase, orientation, diattenuation, and retardance for a variety of calibration samples and biological specimens.
Increasing a microscope's effective field of view via overlapped imaging and machine learning
Oct 10, 2021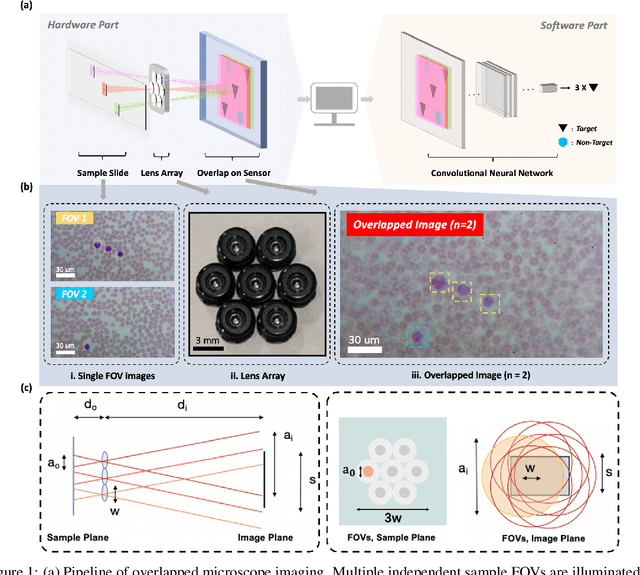
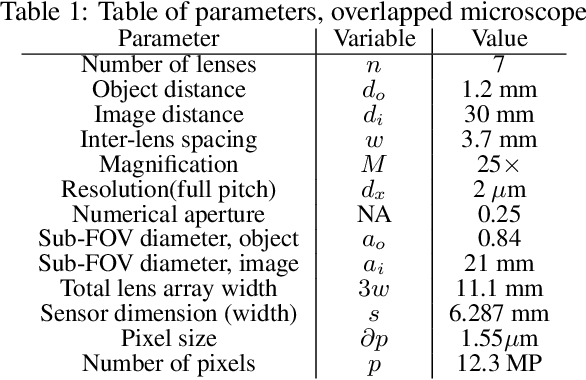
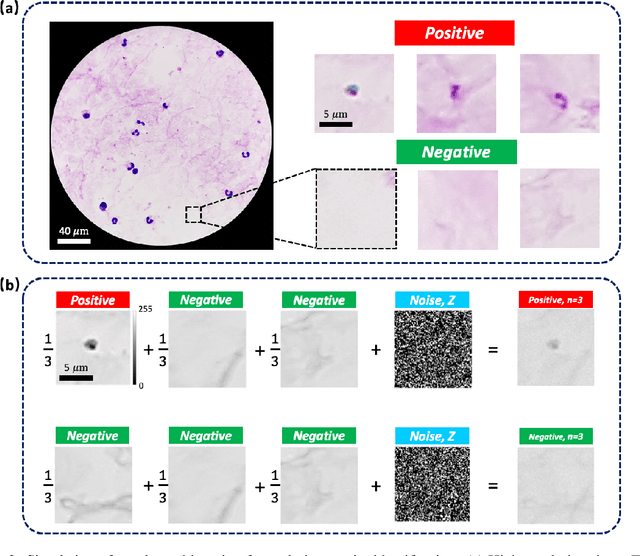
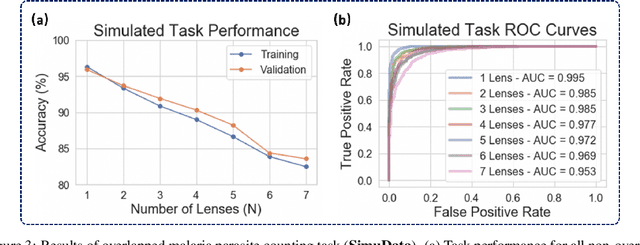
Abstract:This work demonstrates a multi-lens microscopic imaging system that overlaps multiple independent fields of view on a single sensor for high-efficiency automated specimen analysis. Automatic detection, classification and counting of various morphological features of interest is now a crucial component of both biomedical research and disease diagnosis. While convolutional neural networks (CNNs) have dramatically improved the accuracy of counting cells and sub-cellular features from acquired digital image data, the overall throughput is still typically hindered by the limited space-bandwidth product (SBP) of conventional microscopes. Here, we show both in simulation and experiment that overlapped imaging and co-designed analysis software can achieve accurate detection of diagnostically-relevant features for several applications, including counting of white blood cells and the malaria parasite, leading to multi-fold increase in detection and processing throughput with minimal reduction in accuracy.
Imaging dynamics beneath turbid media via parallelized single-photon detection
Jul 22, 2021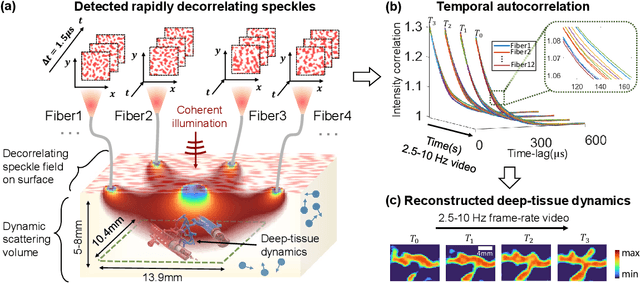
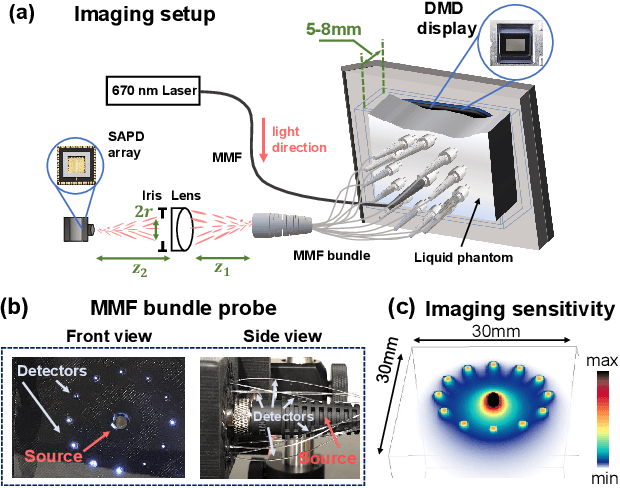
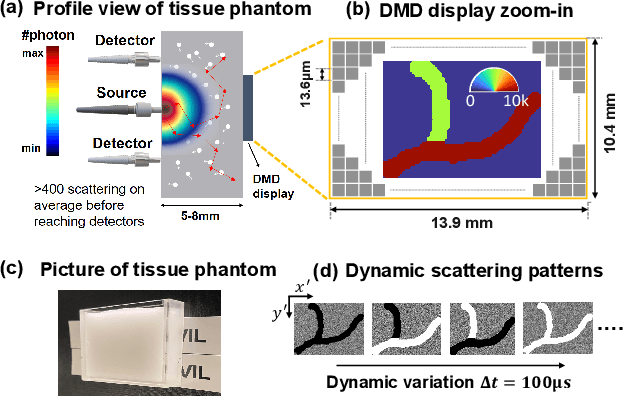
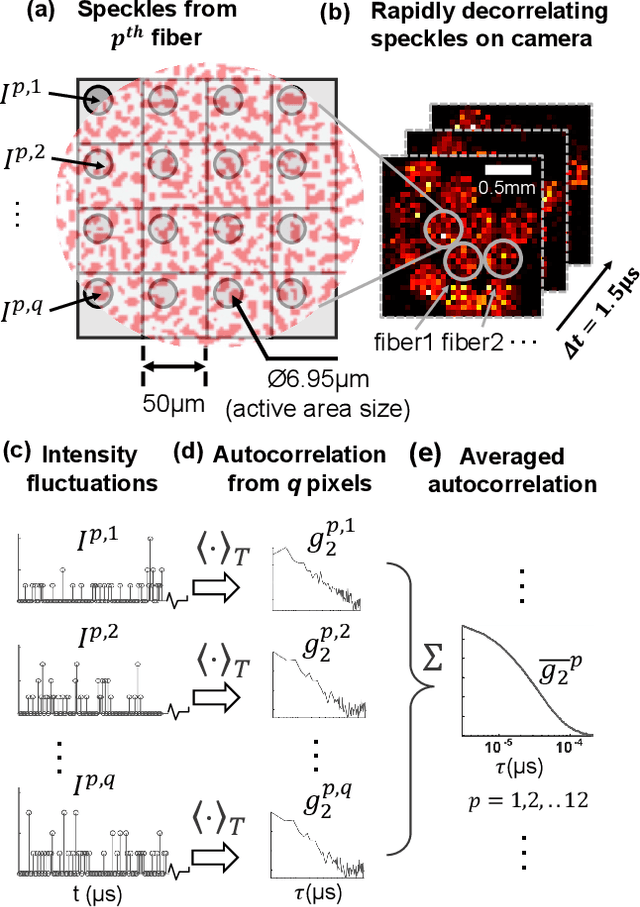
Abstract:Noninvasive optical imaging through dynamic scattering media has numerous important biomedical applications but still remains a challenging task. While standard methods aim to form images based upon optical absorption or fluorescent emission, it is also well-established that the temporal correlation of scattered coherent light diffuses through tissue much like optical intensity. Few works to date, however, have aimed to experimentally measure and process such data to demonstrate deep-tissue imaging of decorrelation dynamics. In this work, we take advantage of a single-photon avalanche diode (SPAD) array camera, with over one thousand detectors, to simultaneously detect speckle fluctuations at the single-photon level from 12 different phantom tissue surface locations delivered via a customized fiber bundle array. We then apply a deep neural network to convert the acquired single-photon measurements into video of scattering dynamics beneath rapidly decorrelating liquid tissue phantoms. We demonstrate the ability to record video of dynamic events occurring 5-8 mm beneath a decorrelating tissue phantom with mm-scale resolution and at a 2.5-10 Hz frame rate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge