Kanghyun Kim
SPLIT: SE(3)-diffusion via Local Geometry-based Score Prediction for 3D Scene-to-Pose-Set Matching Problems
Nov 15, 2024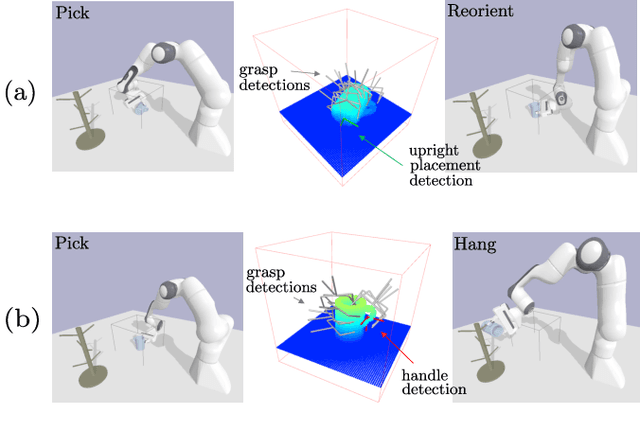
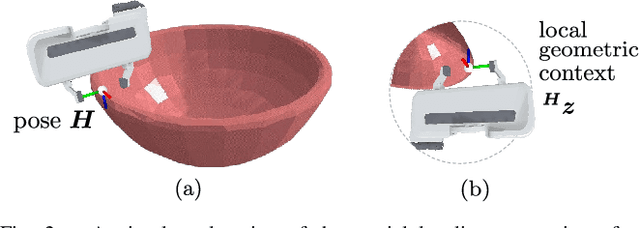
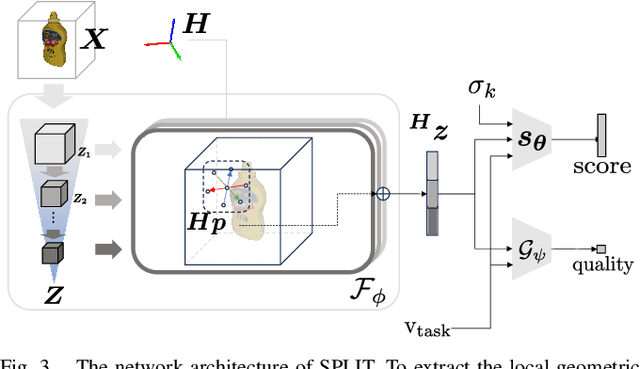
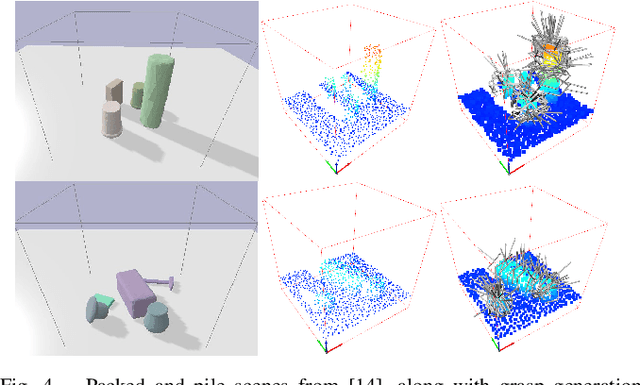
Abstract:To enable versatile robot manipulation, robots must detect task-relevant poses for different purposes from raw scenes. Currently, many perception algorithms are designed for specific purposes, which limits the flexibility of the perception module. We present a general problem formulation called 3D scene-to-pose-set matching, which directly matches the corresponding poses from the scene without relying on task-specific heuristics. To address this, we introduce SPLIT, an SE(3)-diffusion model for generating pose samples from a scene. The model's efficiency comes from predicting scores based on local geometry with respect to the sample pose. Moreover, leveraging the conditioned generation capability of diffusion models, we demonstrate that SPLIT can generate the multi-purpose poses, required to complete both the mug reorientation and hanging manipulation within a single model.
Tensorial tomographic Fourier Ptychography with applications to muscle tissue imaging
May 14, 2023Abstract:We report Tensorial tomographic Fourier Ptychography (ToFu), a new non-scanning label-free tomographic microscopy method for simultaneous imaging of quantitative phase and anisotropic specimen information in 3D. Built upon Fourier Ptychography, a quantitative phase imaging technique, ToFu additionally highlights the vectorial nature of light. The imaging setup consists of a standard microscope equipped with an LED matrix, a polarization generator, and a polarization-sensitive camera. Permittivity tensors of anisotropic samples are computationally recovered from polarized intensity measurements across three dimensions. We demonstrate ToFu's efficiency through volumetric reconstructions of refractive index, birefringence, and orientation for various validation samples, as well as tissue samples from muscle fibers and diseased heart tissue. Our reconstructions of muscle fibers resolve their 3D fine-filament structure and yield consistent morphological measurements compared to gold-standard second harmonic generation scanning confocal microscope images found in the literature. Additionally, we demonstrate reconstructions of a heart tissue sample that carries important polarization information for detecting cardiac amyloidosis.
Digital staining in optical microscopy using deep learning -- a review
Mar 14, 2023Abstract:Until recently, conventional biochemical staining had the undisputed status as well-established benchmark for most biomedical problems related to clinical diagnostics, fundamental research and biotechnology. Despite this role as gold-standard, staining protocols face several challenges, such as a need for extensive, manual processing of samples, substantial time delays, altered tissue homeostasis, limited choice of contrast agents for a given sample, 2D imaging instead of 3D tomography and many more. Label-free optical technologies, on the other hand, do not rely on exogenous and artificial markers, by exploiting intrinsic optical contrast mechanisms, where the specificity is typically less obvious to the human observer. Over the past few years, digital staining has emerged as a promising concept to use modern deep learning for the translation from optical contrast to established biochemical contrast of actual stainings. In this review article, we provide an in-depth analysis of the current state-of-the-art in this field, suggest methods of good practice, identify pitfalls and challenges and postulate promising advances towards potential future implementations and applications.
A Reachability Tree-Based Algorithm for Robot Task and Motion Planning
Mar 07, 2023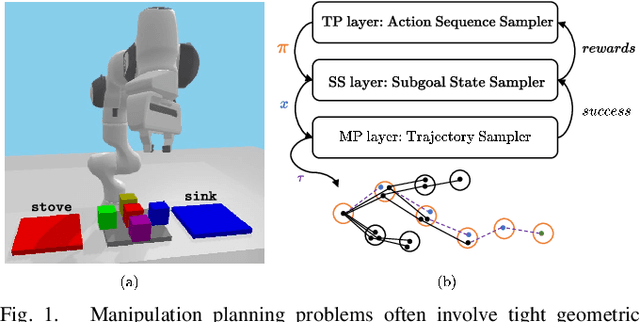
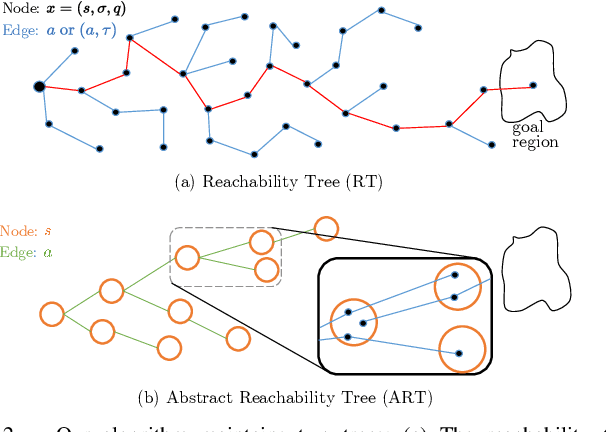
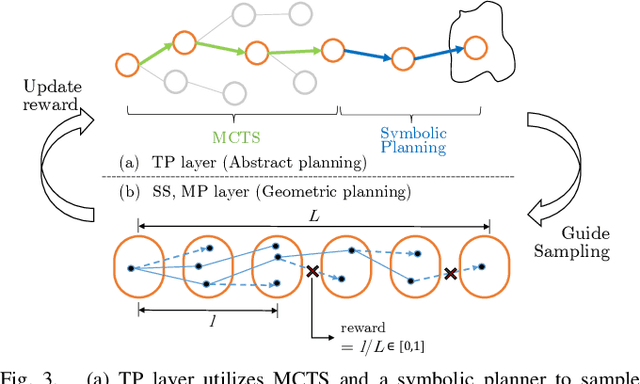
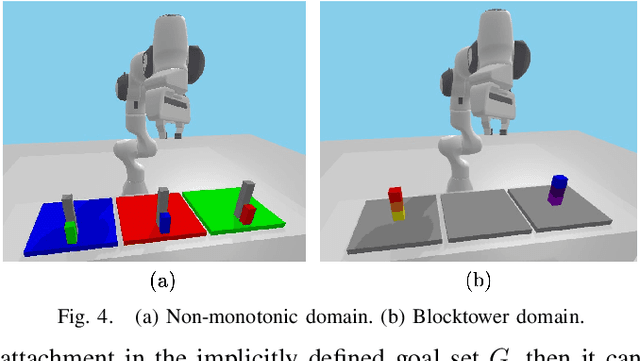
Abstract:This paper presents a novel algorithm for robot task and motion planning (TAMP) problems by utilizing a reachability tree. While tree-based algorithms are known for their speed and simplicity in motion planning (MP), they are not well-suited for TAMP problems that involve both abstracted and geometrical state variables. To address this challenge, we propose a hierarchical sampling strategy, which first generates an abstracted task plan using Monte Carlo tree search (MCTS) and then fills in the details with a geometrically feasible motion trajectory. Moreover, we show that the performance of the proposed method can be significantly enhanced by selecting an appropriate reward for MCTS and by using a pre-generated goal state that is guaranteed to be geometrically feasible. A comparative study using TAMP benchmark problems demonstrates the effectiveness of the proposed approach.
Parallelized computational 3D video microscopy of freely moving organisms at multiple gigapixels per second
Jan 19, 2023Abstract:To study the behavior of freely moving model organisms such as zebrafish (Danio rerio) and fruit flies (Drosophila) across multiple spatial scales, it would be ideal to use a light microscope that can resolve 3D information over a wide field of view (FOV) at high speed and high spatial resolution. However, it is challenging to design an optical instrument to achieve all of these properties simultaneously. Existing techniques for large-FOV microscopic imaging and for 3D image measurement typically require many sequential image snapshots, thus compromising speed and throughput. Here, we present 3D-RAPID, a computational microscope based on a synchronized array of 54 cameras that can capture high-speed 3D topographic videos over a 135-cm^2 area, achieving up to 230 frames per second at throughputs exceeding 5 gigapixels (GPs) per second. 3D-RAPID features a 3D reconstruction algorithm that, for each synchronized temporal snapshot, simultaneously fuses all 54 images seamlessly into a globally-consistent composite that includes a coregistered 3D height map. The self-supervised 3D reconstruction algorithm itself trains a spatiotemporally-compressed convolutional neural network (CNN) that maps raw photometric images to 3D topography, using stereo overlap redundancy and ray-propagation physics as the only supervision mechanism. As a result, our end-to-end 3D reconstruction algorithm is robust to generalization errors and scales to arbitrarily long videos from arbitrarily sized camera arrays. The scalable hardware and software design of 3D-RAPID addresses a longstanding problem in the field of behavioral imaging, enabling parallelized 3D observation of large collections of freely moving organisms at high spatiotemporal throughputs, which we demonstrate in ants (Pogonomyrmex barbatus), fruit flies, and zebrafish larvae.
Multi-scale gigapixel microscopy using a multi-camera array microscope
Nov 30, 2022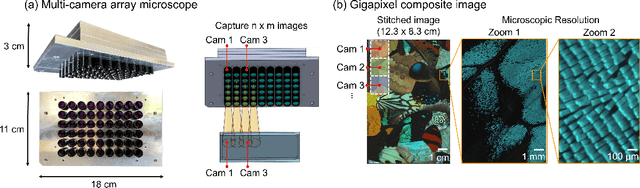
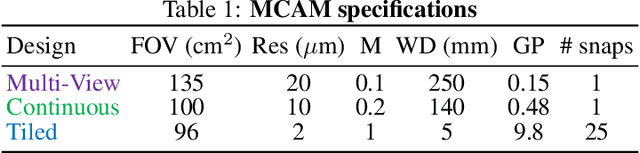
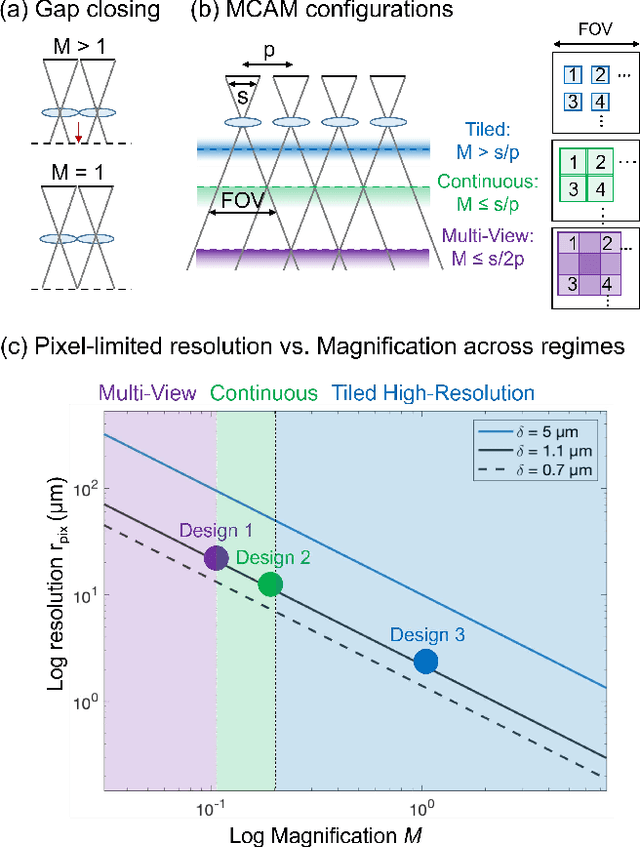
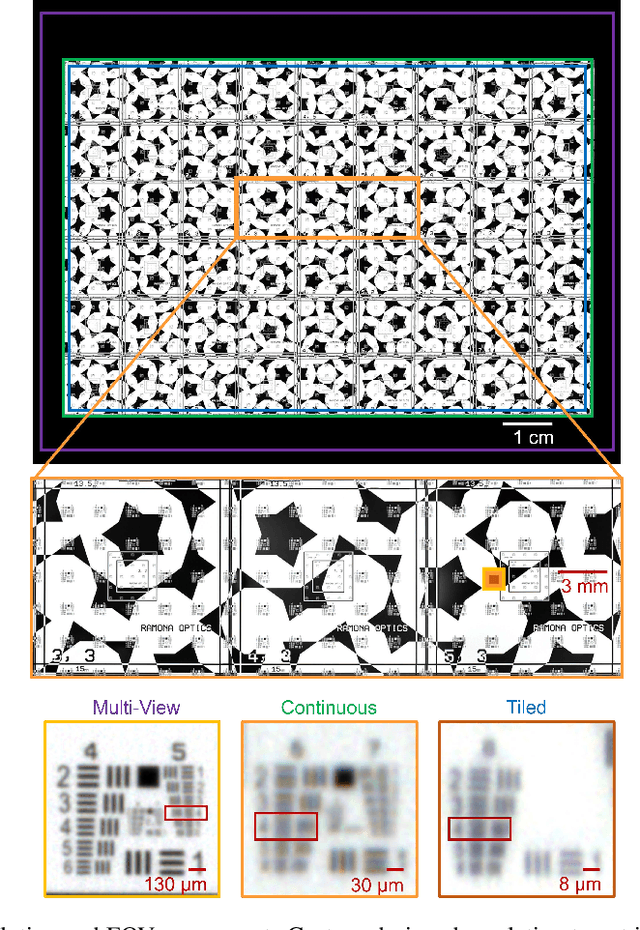
Abstract:This article experimentally examines different configurations of a novel multi-camera array microscope (MCAM) imaging technology. The MCAM is based upon a densely packed array of "micro-cameras" to jointly image across a large field-of-view at high resolution. Each micro-camera within the array images a unique area of a sample of interest, and then all acquired data with 54 micro-cameras are digitally combined into composite frames, whose total pixel counts significantly exceed the pixel counts of standard microscope systems. We present results from three unique MCAM configurations for different use cases. First, we demonstrate a configuration that simultaneously images and estimates the 3D object depth across a 100 x 135 mm^2 field-of-view (FOV) at approximately 20 um resolution, which results in 0.15 gigapixels (GP) per snapshot. Second, we demonstrate an MCAM configuration that records video across a continuous 83 x 123 mm^2 FOV with two-fold increased resolution (0.48 GP per frame). Finally, we report a third high-resolution configuration (2 um resolution) that can rapidly produce 9.8 GP composites of large histopathology specimens.
Tensorial tomographic differential phase-contrast microscopy
Apr 25, 2022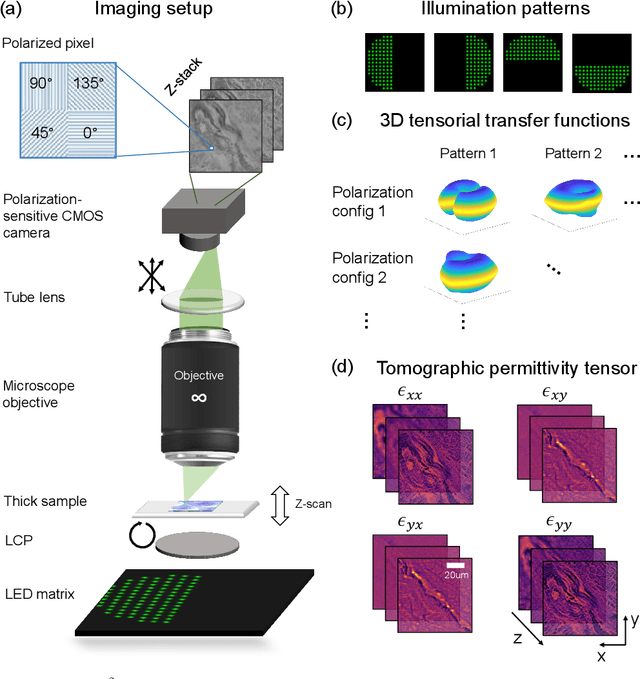
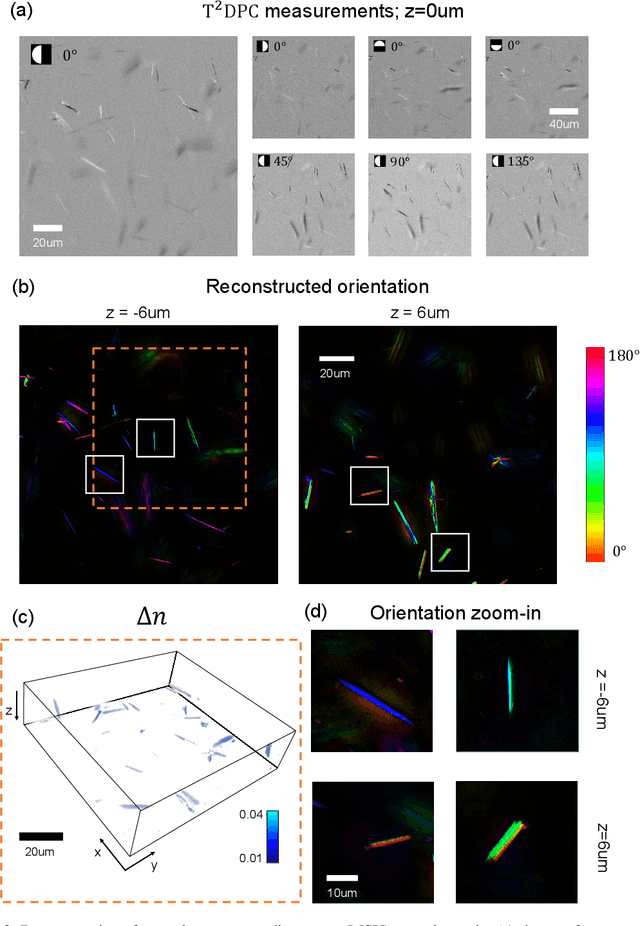
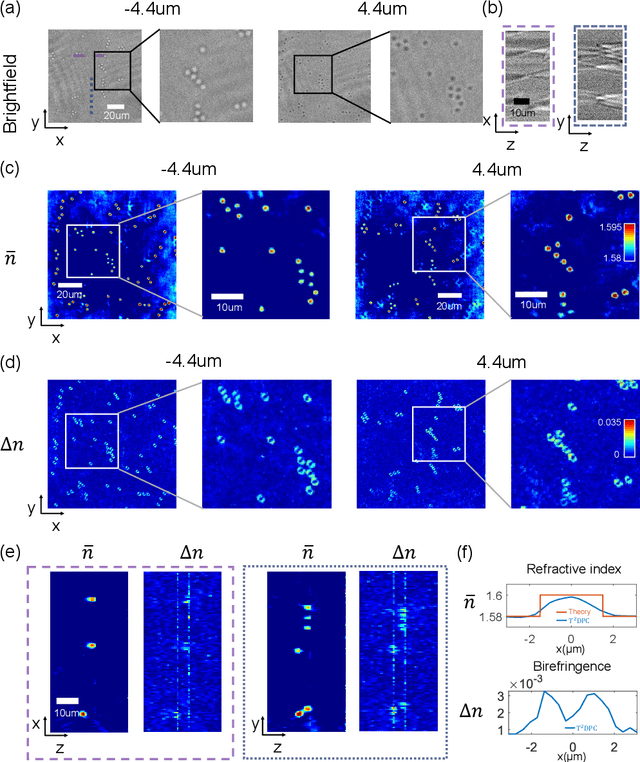
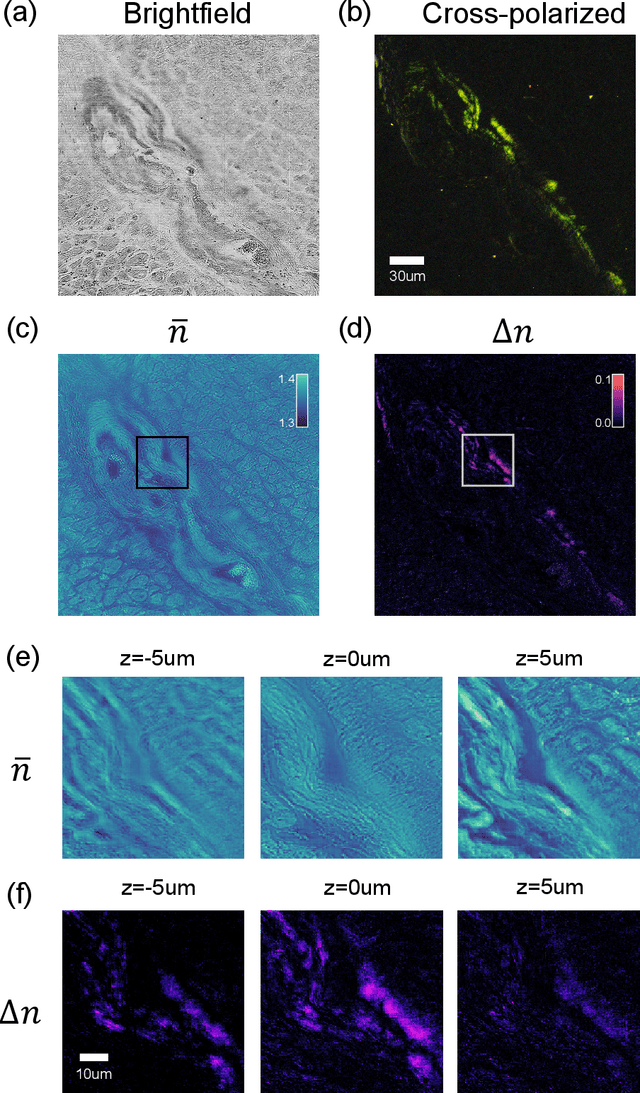
Abstract:We report Tensorial Tomographic Differential Phase-Contrast microscopy (T2DPC), a quantitative label-free tomographic imaging method for simultaneous measurement of phase and anisotropy. T2DPC extends differential phase-contrast microscopy, a quantitative phase imaging technique, to highlight the vectorial nature of light. The method solves for permittivity tensor of anisotropic samples from intensity measurements acquired with a standard microscope equipped with an LED matrix, a circular polarizer, and a polarization-sensitive camera. We demonstrate accurate volumetric reconstructions of refractive index, birefringence, and orientation for various validation samples, and show that the reconstructed polarization structures of a biological specimen are predictive of pathology.
Transient motion classification through turbid volumes via parallelized single-photon detection and deep contrastive embedding
Apr 04, 2022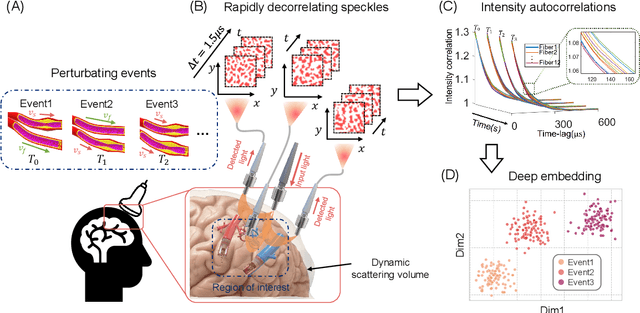
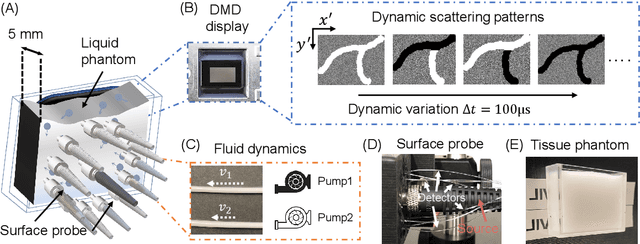
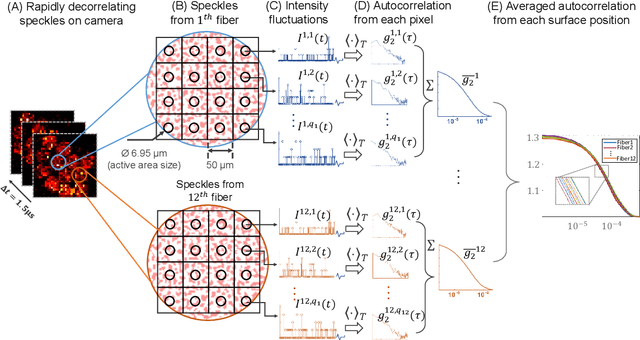
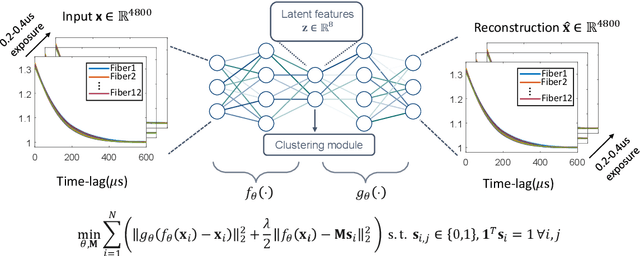
Abstract:Fast noninvasive probing of spatially varying decorrelating events, such as cerebral blood flow beneath the human skull, is an essential task in various scientific and clinical settings. One of the primary optical techniques used is diffuse correlation spectroscopy (DCS), whose classical implementation uses a single or few single-photon detectors, resulting in poor spatial localization accuracy and relatively low temporal resolution. Here, we propose a technique termed Classifying Rapid decorrelation Events via Parallelized single photon dEtection (CREPE)}, a new form of DCS that can probe and classify different decorrelating movements hidden underneath turbid volume with high sensitivity using parallelized speckle detection from a $32\times32$ pixel SPAD array. We evaluate our setup by classifying different spatiotemporal-decorrelating patterns hidden beneath a 5mm tissue-like phantom made with rapidly decorrelating dynamic scattering media. Twelve multi-mode fibers are used to collect scattered light from different positions on the surface of the tissue phantom. To validate our setup, we generate perturbed decorrelation patterns by both a digital micromirror device (DMD) modulated at multi-kilo-hertz rates, as well as a vessel phantom containing flowing fluid. Along with a deep contrastive learning algorithm that outperforms classic unsupervised learning methods, we demonstrate our approach can accurately detect and classify different transient decorrelation events (happening in 0.1-0.4s) underneath turbid scattering media, without any data labeling. This has the potential to be applied to noninvasively monitor deep tissue motion patterns, for example identifying normal or abnormal cerebral blood flow events, at multi-Hertz rates within a compact and static detection probe.
Increasing a microscope's effective field of view via overlapped imaging and machine learning
Oct 10, 2021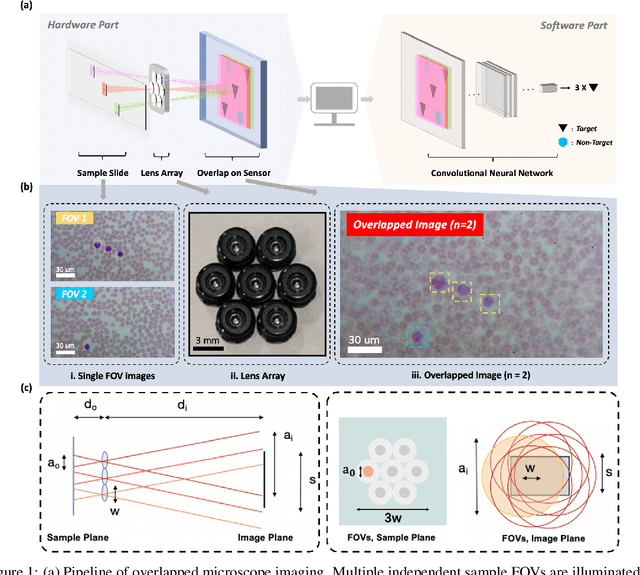
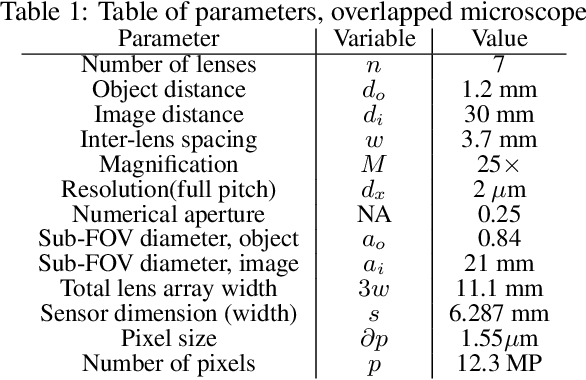
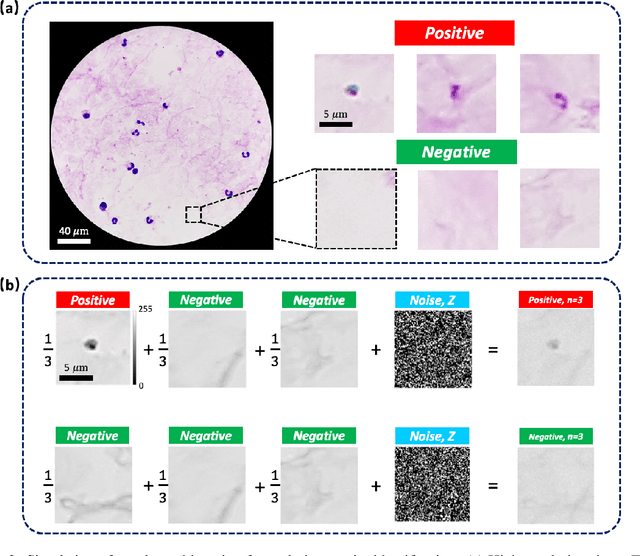
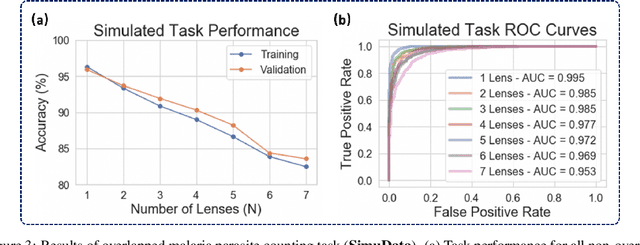
Abstract:This work demonstrates a multi-lens microscopic imaging system that overlaps multiple independent fields of view on a single sensor for high-efficiency automated specimen analysis. Automatic detection, classification and counting of various morphological features of interest is now a crucial component of both biomedical research and disease diagnosis. While convolutional neural networks (CNNs) have dramatically improved the accuracy of counting cells and sub-cellular features from acquired digital image data, the overall throughput is still typically hindered by the limited space-bandwidth product (SBP) of conventional microscopes. Here, we show both in simulation and experiment that overlapped imaging and co-designed analysis software can achieve accurate detection of diagnostically-relevant features for several applications, including counting of white blood cells and the malaria parasite, leading to multi-fold increase in detection and processing throughput with minimal reduction in accuracy.
Multi-element microscope optimization by a learned sensing network with composite physical layers
Jun 27, 2020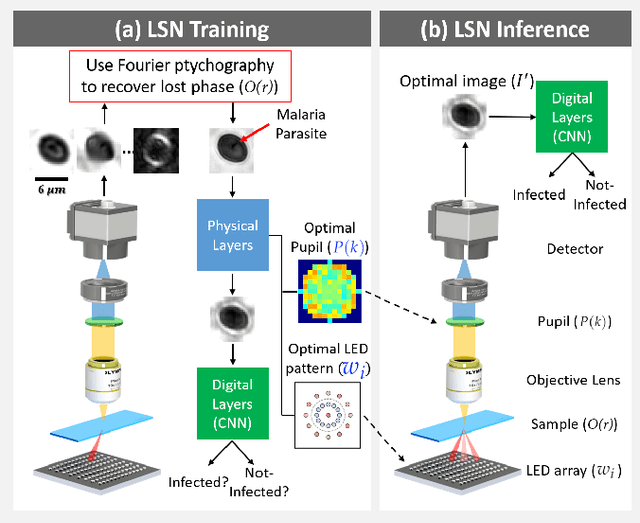


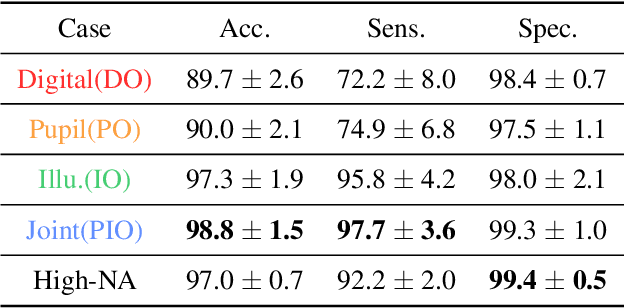
Abstract:Standard microscopes offer a variety of settings to help improve the visibility of different specimens to the end microscope user. Increasingly, however, digital microscopes are used to capture images for automated interpretation by computer algorithms (e.g., for feature classification, detection or segmentation), often without any human involvement. In this work, we investigate an approach to jointly optimize multiple microscope settings, together with a classification network, for improved performance with such automated tasks. We explore the interplay between optimization of programmable illumination and pupil transmission, using experimentally imaged blood smears for automated malaria parasite detection, to show that multi-element "learned sensing" outperforms its single-element counterpart. While not necessarily ideal for human interpretation, the network's resulting low-resolution microscope images (20X-comparable) offer a machine learning network sufficient contrast to match the classification performance of corresponding high-resolution imagery (100X-comparable), pointing a path towards accurate automation over large fields-of-view.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge