Lucas Kreiss
Detecting immune cells with label-free two-photon autofluorescence and deep learning
Jun 17, 2025Abstract:Label-free imaging has gained broad interest because of its potential to omit elaborate staining procedures which is especially relevant for in vivo use. Label-free multiphoton microscopy (MPM), for instance, exploits two-photon excitation of natural autofluorescence (AF) from native, metabolic proteins, making it ideal for in vivo endomicroscopy. Deep learning (DL) models have been widely used in other optical imaging technologies to predict specific target annotations and thereby digitally augment the specificity of these label-free images. However, this computational specificity has only rarely been implemented for MPM. In this work, we used a data set of label-free MPM images from a series of different immune cell types (5,075 individual cells for binary classification in mixed samples and 3,424 cells for a multi-class classification task) and trained a convolutional neural network (CNN) to classify cell types based on this label-free AF as input. A low-complexity squeezeNet architecture was able to achieve reliable immune cell classification results (0.89 ROC-AUC, 0.95 PR-AUC, for binary classification in mixed samples; 0.689 F1 score, 0.697 precision, 0.748 recall, and 0.683 MCC for six-class classification in isolated samples). Perturbation tests confirmed that the model is not confused by extracellular environment and that both input AF channels (NADH and FAD) are about equally important to the classification. In the future, such predictive DL models could directly detect specific immune cells in unstained images and thus, computationally improve the specificity of label-free MPM which would have great potential for in vivo endomicroscopy.
Tensorial tomographic Fourier Ptychography with applications to muscle tissue imaging
May 14, 2023Abstract:We report Tensorial tomographic Fourier Ptychography (ToFu), a new non-scanning label-free tomographic microscopy method for simultaneous imaging of quantitative phase and anisotropic specimen information in 3D. Built upon Fourier Ptychography, a quantitative phase imaging technique, ToFu additionally highlights the vectorial nature of light. The imaging setup consists of a standard microscope equipped with an LED matrix, a polarization generator, and a polarization-sensitive camera. Permittivity tensors of anisotropic samples are computationally recovered from polarized intensity measurements across three dimensions. We demonstrate ToFu's efficiency through volumetric reconstructions of refractive index, birefringence, and orientation for various validation samples, as well as tissue samples from muscle fibers and diseased heart tissue. Our reconstructions of muscle fibers resolve their 3D fine-filament structure and yield consistent morphological measurements compared to gold-standard second harmonic generation scanning confocal microscope images found in the literature. Additionally, we demonstrate reconstructions of a heart tissue sample that carries important polarization information for detecting cardiac amyloidosis.
Digital staining in optical microscopy using deep learning -- a review
Mar 14, 2023Abstract:Until recently, conventional biochemical staining had the undisputed status as well-established benchmark for most biomedical problems related to clinical diagnostics, fundamental research and biotechnology. Despite this role as gold-standard, staining protocols face several challenges, such as a need for extensive, manual processing of samples, substantial time delays, altered tissue homeostasis, limited choice of contrast agents for a given sample, 2D imaging instead of 3D tomography and many more. Label-free optical technologies, on the other hand, do not rely on exogenous and artificial markers, by exploiting intrinsic optical contrast mechanisms, where the specificity is typically less obvious to the human observer. Over the past few years, digital staining has emerged as a promising concept to use modern deep learning for the translation from optical contrast to established biochemical contrast of actual stainings. In this review article, we provide an in-depth analysis of the current state-of-the-art in this field, suggest methods of good practice, identify pitfalls and challenges and postulate promising advances towards potential future implementations and applications.
Parallelized computational 3D video microscopy of freely moving organisms at multiple gigapixels per second
Jan 19, 2023Abstract:To study the behavior of freely moving model organisms such as zebrafish (Danio rerio) and fruit flies (Drosophila) across multiple spatial scales, it would be ideal to use a light microscope that can resolve 3D information over a wide field of view (FOV) at high speed and high spatial resolution. However, it is challenging to design an optical instrument to achieve all of these properties simultaneously. Existing techniques for large-FOV microscopic imaging and for 3D image measurement typically require many sequential image snapshots, thus compromising speed and throughput. Here, we present 3D-RAPID, a computational microscope based on a synchronized array of 54 cameras that can capture high-speed 3D topographic videos over a 135-cm^2 area, achieving up to 230 frames per second at throughputs exceeding 5 gigapixels (GPs) per second. 3D-RAPID features a 3D reconstruction algorithm that, for each synchronized temporal snapshot, simultaneously fuses all 54 images seamlessly into a globally-consistent composite that includes a coregistered 3D height map. The self-supervised 3D reconstruction algorithm itself trains a spatiotemporally-compressed convolutional neural network (CNN) that maps raw photometric images to 3D topography, using stereo overlap redundancy and ray-propagation physics as the only supervision mechanism. As a result, our end-to-end 3D reconstruction algorithm is robust to generalization errors and scales to arbitrarily long videos from arbitrarily sized camera arrays. The scalable hardware and software design of 3D-RAPID addresses a longstanding problem in the field of behavioral imaging, enabling parallelized 3D observation of large collections of freely moving organisms at high spatiotemporal throughputs, which we demonstrate in ants (Pogonomyrmex barbatus), fruit flies, and zebrafish larvae.
Multi-scale gigapixel microscopy using a multi-camera array microscope
Nov 30, 2022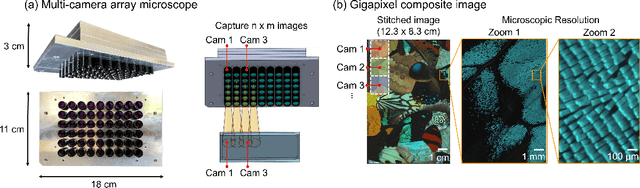
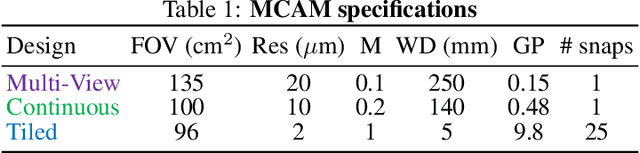
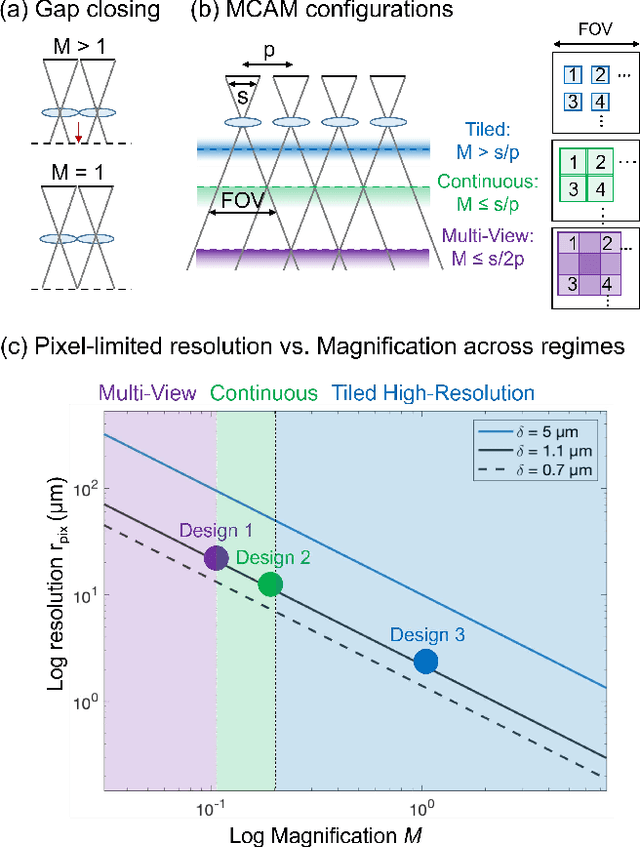
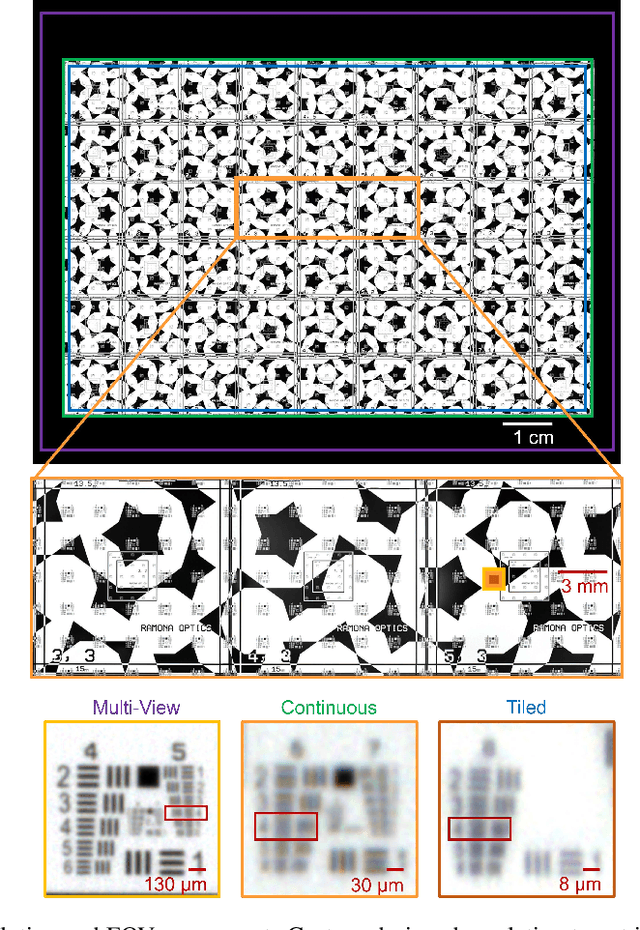
Abstract:This article experimentally examines different configurations of a novel multi-camera array microscope (MCAM) imaging technology. The MCAM is based upon a densely packed array of "micro-cameras" to jointly image across a large field-of-view at high resolution. Each micro-camera within the array images a unique area of a sample of interest, and then all acquired data with 54 micro-cameras are digitally combined into composite frames, whose total pixel counts significantly exceed the pixel counts of standard microscope systems. We present results from three unique MCAM configurations for different use cases. First, we demonstrate a configuration that simultaneously images and estimates the 3D object depth across a 100 x 135 mm^2 field-of-view (FOV) at approximately 20 um resolution, which results in 0.15 gigapixels (GP) per snapshot. Second, we demonstrate an MCAM configuration that records video across a continuous 83 x 123 mm^2 FOV with two-fold increased resolution (0.48 GP per frame). Finally, we report a third high-resolution configuration (2 um resolution) that can rapidly produce 9.8 GP composites of large histopathology specimens.
SEMPAI: a Self-Enhancing Multi-Photon Artificial Intelligence for prior-informed assessment of muscle function and pathology
Oct 28, 2022Abstract:Deep learning (DL) shows notable success in biomedical studies. However, most DL algorithms work as a black box, exclude biomedical experts, and need extensive data. We introduce the Self-Enhancing Multi-Photon Artificial Intelligence (SEMPAI), that integrates hypothesis-driven priors in a data-driven DL approach for research on multiphoton microscopy (MPM) of muscle fibers. SEMPAI utilizes meta-learning to optimize prior integration, data representation, and neural network architecture simultaneously. This allows hypothesis testing and provides interpretable feedback about the origin of biological information in MPM images. SEMPAI performs joint learning of several tasks to enable prediction for small datasets. The method is applied on an extensive multi-study dataset resulting in the largest joint analysis of pathologies and function for single muscle fibers. SEMPAI outperforms state-of-the-art biomarkers in six of seven predictive tasks, including those with scarce data. SEMPAI's DL models with integrated priors are superior to those without priors and to prior-only machine learning approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge