Maxime W. Lafarge
Domain generalization across tumor types, laboratories, and species -- insights from the 2022 edition of the Mitosis Domain Generalization Challenge
Sep 27, 2023



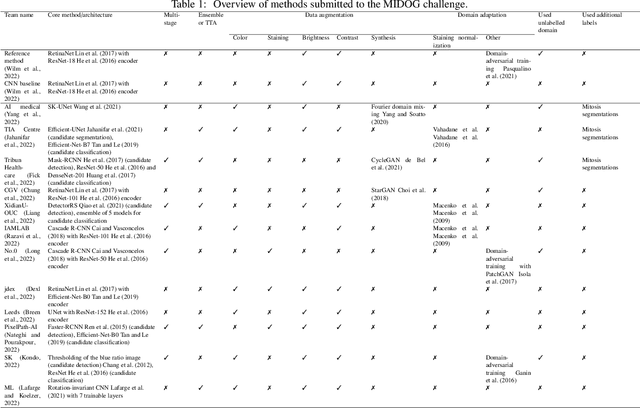
Abstract:Recognition of mitotic figures in histologic tumor specimens is highly relevant to patient outcome assessment. This task is challenging for algorithms and human experts alike, with deterioration of algorithmic performance under shifts in image representations. Considerable covariate shifts occur when assessment is performed on different tumor types, images are acquired using different digitization devices, or specimens are produced in different laboratories. This observation motivated the inception of the 2022 challenge on MItosis Domain Generalization (MIDOG 2022). The challenge provided annotated histologic tumor images from six different domains and evaluated the algorithmic approaches for mitotic figure detection provided by nine challenge participants on ten independent domains. Ground truth for mitotic figure detection was established in two ways: a three-expert consensus and an independent, immunohistochemistry-assisted set of labels. This work represents an overview of the challenge tasks, the algorithmic strategies employed by the participants, and potential factors contributing to their success. With an $F_1$ score of 0.764 for the top-performing team, we summarize that domain generalization across various tumor domains is possible with today's deep learning-based recognition pipelines. When assessed against the immunohistochemistry-assisted reference standard, all methods resulted in reduced recall scores, but with only minor changes in the order of participants in the ranking.
CohortFinder: an open-source tool for data-driven partitioning of biomedical image cohorts to yield robust machine learning models
Jul 17, 2023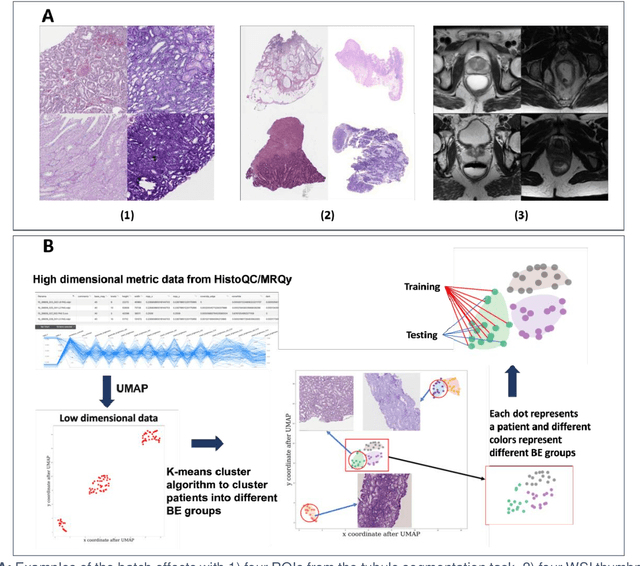

Abstract:Batch effects (BEs) refer to systematic technical differences in data collection unrelated to biological variations whose noise is shown to negatively impact machine learning (ML) model generalizability. Here we release CohortFinder, an open-source tool aimed at mitigating BEs via data-driven cohort partitioning. We demonstrate CohortFinder improves ML model performance in downstream medical image processing tasks. CohortFinder is freely available for download at cohortfinder.com.
Multi-task learning for tissue segmentation and tumor detection in colorectal cancer histology slides
Apr 06, 2023



Abstract:Automating tissue segmentation and tumor detection in histopathology images of colorectal cancer (CRC) is an enabler for faster diagnostic pathology workflows. At the same time it is a challenging task due to low availability of public annotated datasets and high variability of image appearance. The semi-supervised learning for CRC detection (SemiCOL) challenge 2023 provides partially annotated data to encourage the development of automated solutions for tissue segmentation and tumor detection. We propose a U-Net based multi-task model combined with channel-wise and image-statistics-based color augmentations, as well as test-time augmentation, as a candidate solution to the SemiCOL challenge. Our approach achieved a multi-task Dice score of .8655 (Arm 1) and .8515 (Arm 2) for tissue segmentation and AUROC of .9725 (Arm 1) and 0.9750 (Arm 2) for tumor detection on the challenge validation set. The source code for our approach is made publicly available at https://github.com/lely475/CTPLab_SemiCOL2023.
Fine-Grained Hard Negative Mining: Generalizing Mitosis Detection with a Fifth of the MIDOG 2022 Dataset
Jan 03, 2023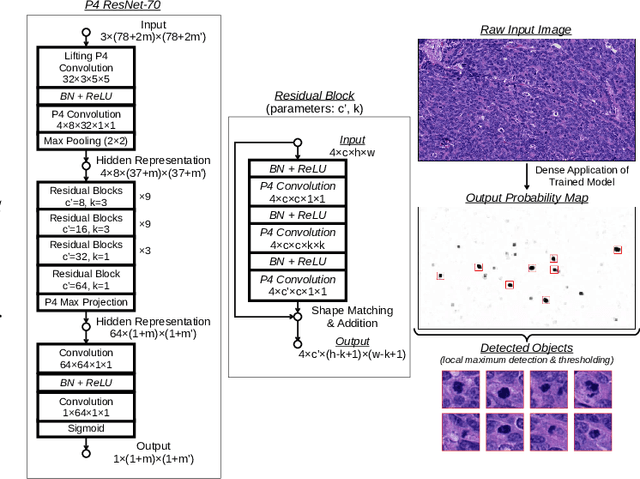
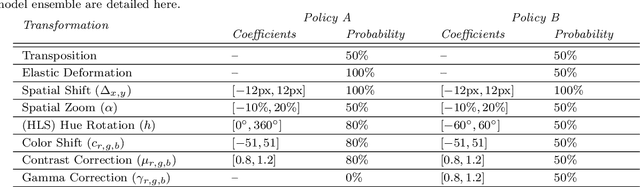
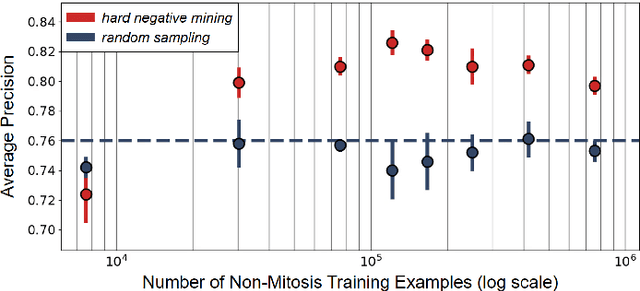
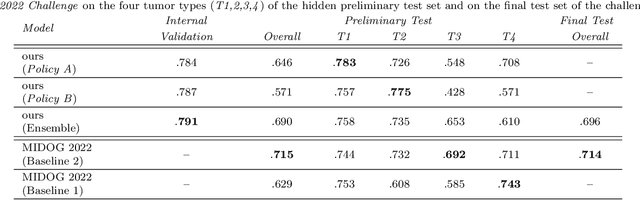
Abstract:Making histopathology image classifiers robust to a wide range of real-world variability is a challenging task. Here, we describe a candidate deep learning solution for the Mitosis Domain Generalization Challenge 2022 (MIDOG) to address the problem of generalization for mitosis detection in images of hematoxylin-eosin-stained histology slides under high variability (scanner, tissue type and species variability). Our approach consists in training a rotation-invariant deep learning model using aggressive data augmentation with a training set enriched with hard negative examples and automatically selected negative examples from the unlabeled part of the challenge dataset. To optimize the performance of our models, we investigated a hard negative mining regime search procedure that lead us to train our best model using a subset of image patches representing 19.6% of our training partition of the challenge dataset. Our candidate model ensemble achieved a F1-score of .697 on the final test set after automated evaluation on the challenge platform, achieving the third best overall score in the MIDOG 2022 Challenge.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022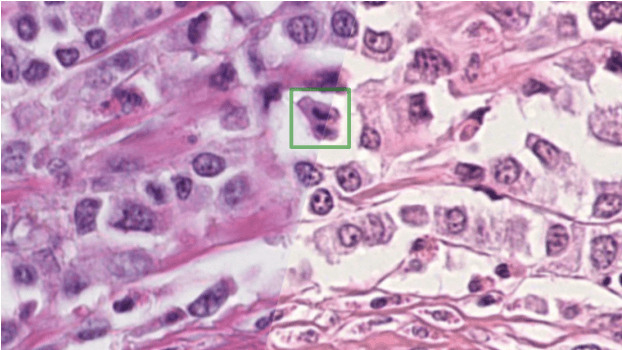
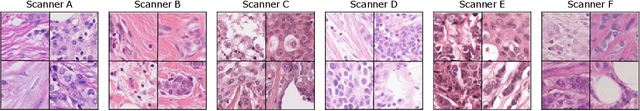
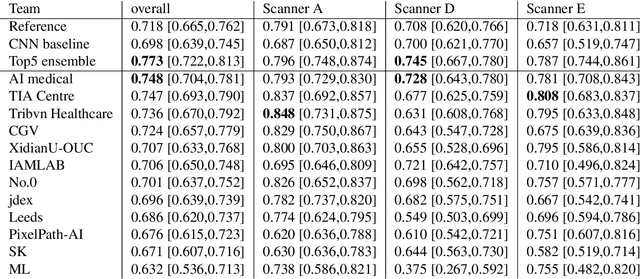
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
Rotation Invariance and Extensive Data Augmentation: a strategy for the Mitosis Domain Generalization (MIDOG) Challenge
Sep 26, 2021



Abstract:Automated detection of mitotic figures in histopathology images is a challenging task: here, we present the different steps that describe the strategy we applied to participate in the MIDOG 2021 competition. The purpose of the competition was to evaluate the generalization of solutions to images acquired with unseen target scanners (hidden for the participants) under the constraint of using training data from a limited set of four independent source scanners. Given this goal and constraints, we joined the challenge by proposing a straight-forward solution based on a combination of state-of-the-art deep learning methods with the aim of yielding robustness to possible scanner-related distributional shifts at inference time. Our solution combines methods that were previously shown to be efficient for mitosis detection: hard negative mining, extensive data augmentation, rotation-invariant convolutional networks. We trained five models with different splits of the provided dataset. The subsequent classifiers produced F1-scores with a mean and standard deviation of 0.747+/-0.032 on the test splits. The resulting ensemble constitutes our candidate algorithm: its automated evaluation on the preliminary test set of the challenge returned a F1-score of 0.6828.
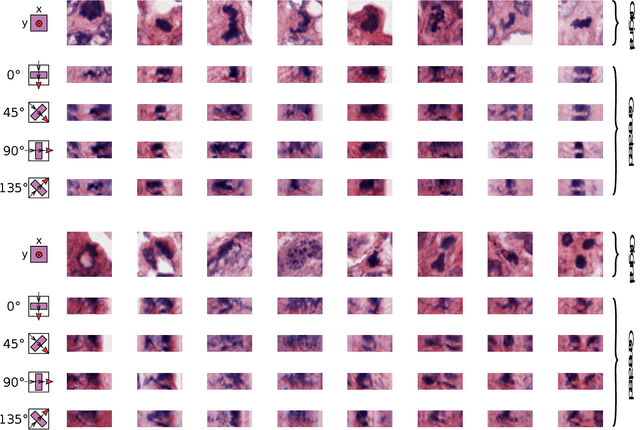
Orientation-Disentangled Unsupervised Representation Learning for Computational Pathology
Aug 26, 2020



Abstract:Unsupervised learning enables modeling complex images without the need for annotations. The representation learned by such models can facilitate any subsequent analysis of large image datasets. However, some generative factors that cause irrelevant variations in images can potentially get entangled in such a learned representation causing the risk of negatively affecting any subsequent use. The orientation of imaged objects, for instance, is often arbitrary/irrelevant, thus it can be desired to learn a representation in which the orientation information is disentangled from all other factors. Here, we propose to extend the Variational Auto-Encoder framework by leveraging the group structure of rotation-equivariant convolutional networks to learn orientation-wise disentangled generative factors of histopathology images. This way, we enforce a novel partitioning of the latent space, such that oriented and isotropic components get separated. We evaluated this structured representation on a dataset that consists of tissue regions for which nuclear pleomorphism and mitotic activity was assessed by expert pathologists. We show that the trained models efficiently disentangle the inherent orientation information of single-cell images. In comparison to classical approaches, the resulting aggregated representation of sub-populations of cells produces higher performances in subsequent tasks.
Roto-Translation Equivariant Convolutional Networks: Application to Histopathology Image Analysis
Feb 20, 2020



Abstract:Rotation-invariance is a desired property of machine-learning models for medical image analysis and in particular for computational pathology applications. We propose a framework to encode the geometric structure of the special Euclidean motion group SE(2) in convolutional networks to yield translation and rotation equivariance via the introduction of SE(2)-group convolution layers. This structure enables models to learn feature representations with a discretized orientation dimension that guarantees that their outputs are invariant under a discrete set of rotations. Conventional approaches for rotation invariance rely mostly on data augmentation, but this does not guarantee the robustness of the output when the input is rotated. At that, trained conventional CNNs may require test-time rotation augmentation to reach their full capability. This study is focused on histopathology image analysis applications for which it is desirable that the arbitrary global orientation information of the imaged tissues is not captured by the machine learning models. The proposed framework is evaluated on three different histopathology image analysis tasks (mitosis detection, nuclei segmentation and tumor classification). We present a comparative analysis for each problem and show that consistent increase of performances can be achieved when using the proposed framework.
Inferring a Third Spatial Dimension from 2D Histological Images
Jan 10, 2018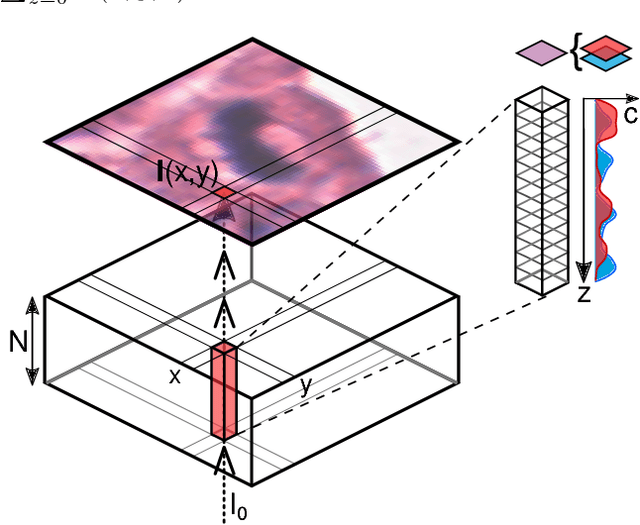
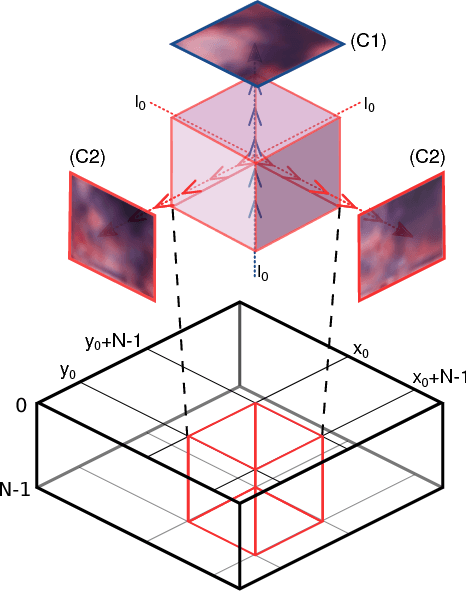
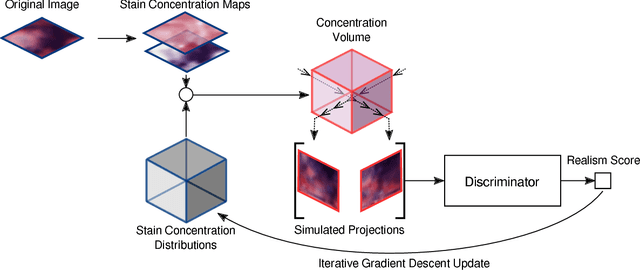
Abstract:Histological images are obtained by transmitting light through a tissue specimen that has been stained in order to produce contrast. This process results in 2D images of the specimen that has a three-dimensional structure. In this paper, we propose a method to infer how the stains are distributed in the direction perpendicular to the surface of the slide for a given 2D image in order to obtain a 3D representation of the tissue. This inference is achieved by decomposition of the staining concentration maps under constraints that ensure realistic decomposition and reconstruction of the original 2D images. Our study shows that it is possible to generate realistic 3D images making this method a potential tool for data augmentation when training deep learning models.
Domain-adversarial neural networks to address the appearance variability of histopathology images
Jul 19, 2017



Abstract:Preparing and scanning histopathology slides consists of several steps, each with a multitude of parameters. The parameters can vary between pathology labs and within the same lab over time, resulting in significant variability of the tissue appearance that hampers the generalization of automatic image analysis methods. Typically, this is addressed with ad-hoc approaches such as staining normalization that aim to reduce the appearance variability. In this paper, we propose a systematic solution based on domain-adversarial neural networks. We hypothesize that removing the domain information from the model representation leads to better generalization. We tested our hypothesis for the problem of mitosis detection in breast cancer histopathology images and made a comparative analysis with two other approaches. We show that combining color augmentation with domain-adversarial training is a better alternative than standard approaches to improve the generalization of deep learning methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge