Lydia A. Schoenpflug
Revisiting Automatic Data Curation for Vision Foundation Models in Digital Pathology
Mar 24, 2025Abstract:Vision foundation models (FMs) are accelerating the development of digital pathology algorithms and transforming biomedical research. These models learn, in a self-supervised manner, to represent histological features in highly heterogeneous tiles extracted from whole-slide images (WSIs) of real-world patient samples. The performance of these FMs is significantly influenced by the size, diversity, and balance of the pre-training data. However, data selection has been primarily guided by expert knowledge at the WSI level, focusing on factors such as disease classification and tissue types, while largely overlooking the granular details available at the tile level. In this paper, we investigate the potential of unsupervised automatic data curation at the tile-level, taking into account 350 million tiles. Specifically, we apply hierarchical clustering trees to pre-extracted tile embeddings, allowing us to sample balanced datasets uniformly across the embedding space of the pretrained FM. We further identify these datasets are subject to a trade-off between size and balance, potentially compromising the quality of representations learned by FMs, and propose tailored batch sampling strategies to mitigate this effect. We demonstrate the effectiveness of our method through improved performance on a diverse range of clinically relevant downstream tasks.
SoftCTM: Cell detection by soft instance segmentation and consideration of cell-tissue interaction
Dec 19, 2023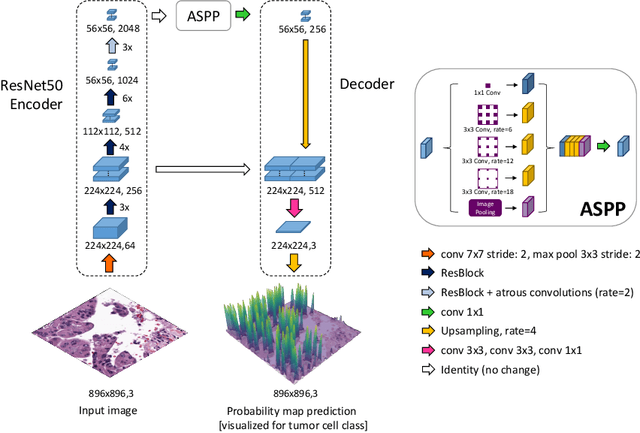
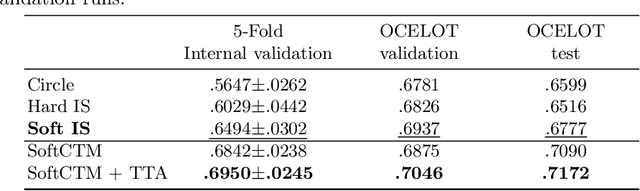
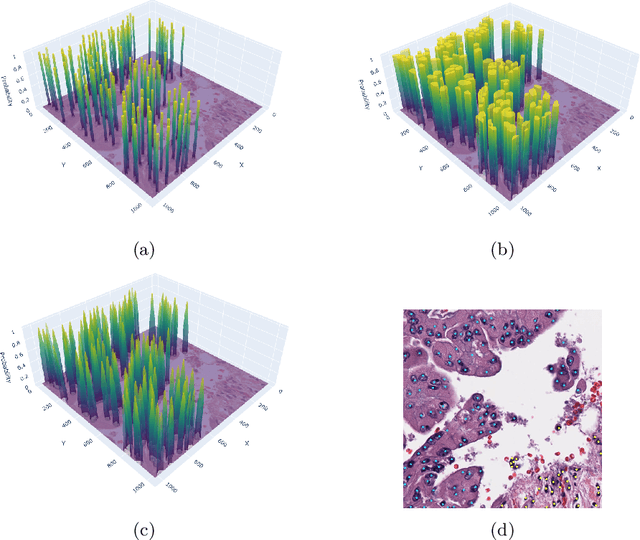

Abstract:Detecting and classifying cells in histopathology H\&E stained whole-slide images is a core task in computational pathology, as it provides valuable insight into the tumor microenvironment. In this work we investigate the impact of ground truth formats on the models performance. Additionally, cell-tissue interactions are considered by providing tissue segmentation predictions as input to the cell detection model. We find that a "soft", probability-map instance segmentation ground truth leads to best model performance. Combined with cell-tissue interaction and test-time augmentation our Soft Cell-Tissue-Model (SoftCTM) achieves 0.7172 mean F1-Score on the Overlapped Cell On Tissue (OCELOT) test set, achieving the third best overall score in the OCELOT 2023 Challenge. The source code for our approach is made publicly available at https://github.com/lely475/ocelot23algo.
Multi-task learning for tissue segmentation and tumor detection in colorectal cancer histology slides
Apr 06, 2023



Abstract:Automating tissue segmentation and tumor detection in histopathology images of colorectal cancer (CRC) is an enabler for faster diagnostic pathology workflows. At the same time it is a challenging task due to low availability of public annotated datasets and high variability of image appearance. The semi-supervised learning for CRC detection (SemiCOL) challenge 2023 provides partially annotated data to encourage the development of automated solutions for tissue segmentation and tumor detection. We propose a U-Net based multi-task model combined with channel-wise and image-statistics-based color augmentations, as well as test-time augmentation, as a candidate solution to the SemiCOL challenge. Our approach achieved a multi-task Dice score of .8655 (Arm 1) and .8515 (Arm 2) for tissue segmentation and AUROC of .9725 (Arm 1) and 0.9750 (Arm 2) for tumor detection on the challenge validation set. The source code for our approach is made publicly available at https://github.com/lely475/CTPLab_SemiCOL2023.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge