Zhiwei Zhai
SELMA3D challenge: Self-supervised learning for 3D light-sheet microscopy image segmentation
Jan 07, 2025Abstract:Recent innovations in light sheet microscopy, paired with developments in tissue clearing techniques, enable the 3D imaging of large mammalian tissues with cellular resolution. Combined with the progress in large-scale data analysis, driven by deep learning, these innovations empower researchers to rapidly investigate the morphological and functional properties of diverse biological samples. Segmentation, a crucial preliminary step in the analysis process, can be automated using domain-specific deep learning models with expert-level performance. However, these models exhibit high sensitivity to domain shifts, leading to a significant drop in accuracy when applied to data outside their training distribution. To address this limitation, and inspired by the recent success of self-supervised learning in training generalizable models, we organized the SELMA3D Challenge during the MICCAI 2024 conference. SELMA3D provides a vast collection of light-sheet images from cleared mice and human brains, comprising 35 large 3D images-each with over 1000^3 voxels-and 315 annotated small patches for finetuning, preliminary testing and final testing. The dataset encompasses diverse biological structures, including vessel-like and spot-like structures. Five teams participated in all phases of the challenge, and their proposed methods are reviewed in this paper. Quantitative and qualitative results from most participating teams demonstrate that self-supervised learning on large datasets improves segmentation model performance and generalization. We will continue to support and extend SELMA3D as an inaugural MICCAI challenge focused on self-supervised learning for 3D microscopy image segmentation.
Olive Branch Learning: A Topology-Aware Federated Learning Framework for Space-Air-Ground Integrated Network
Dec 02, 2022Abstract:The space-air-ground integrated network (SAGIN), one of the key technologies for next-generation mobile communication systems, can facilitate data transmission for users all over the world, especially in some remote areas where vast amounts of informative data are collected by Internet of remote things (IoRT) devices to support various data-driven artificial intelligence (AI) services. However, training AI models centrally with the assistance of SAGIN faces the challenges of highly constrained network topology, inefficient data transmission, and privacy issues. To tackle these challenges, we first propose a novel topology-aware federated learning framework for the SAGIN, namely Olive Branch Learning (OBL). Specifically, the IoRT devices in the ground layer leverage their private data to perform model training locally, while the air nodes in the air layer and the ring-structured low earth orbit (LEO) satellite constellation in the space layer are in charge of model aggregation (synchronization) at different scales.To further enhance communication efficiency and inference performance of OBL, an efficient Communication and Non-IID-aware Air node-Satellite Assignment (CNASA) algorithm is designed by taking the data class distribution of the air nodes as well as their geographic locations into account. Furthermore, we extend our OBL framework and CNASA algorithm to adapt to more complex multi-orbit satellite networks. We analyze the convergence of our OBL framework and conclude that the CNASA algorithm contributes to the fast convergence of the global model. Extensive experiments based on realistic datasets corroborate the superior performance of our algorithm over the benchmark policies.
Multi-task Semi-supervised Learning for Pulmonary Lobe Segmentation
Apr 22, 2021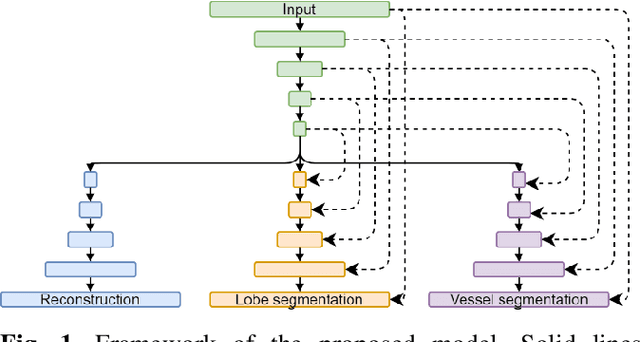
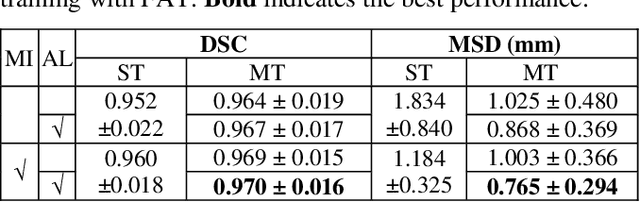
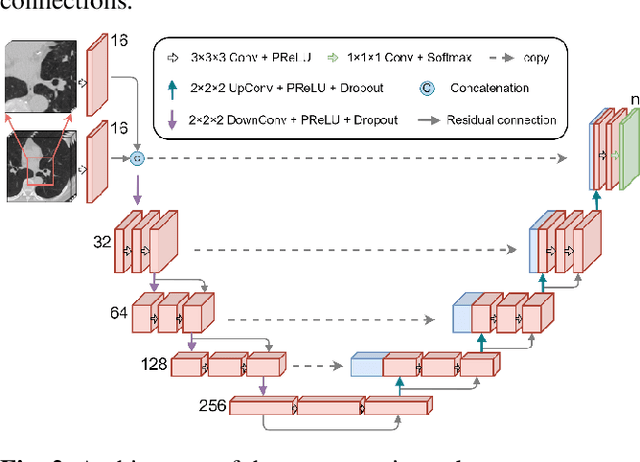
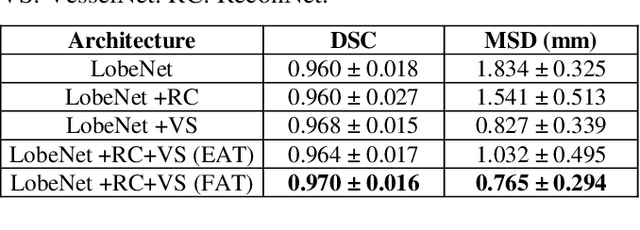
Abstract:Pulmonary lobe segmentation is an important preprocessing task for the analysis of lung diseases. Traditional methods relying on fissure detection or other anatomical features, such as the distribution of pulmonary vessels and airways, could provide reasonably accurate lobe segmentations. Deep learning based methods can outperform these traditional approaches, but require large datasets. Deep multi-task learning is expected to utilize labels of multiple different structures. However, commonly such labels are distributed over multiple datasets. In this paper, we proposed a multi-task semi-supervised model that can leverage information of multiple structures from unannotated datasets and datasets annotated with different structures. A focused alternating training strategy is presented to balance the different tasks. We evaluated the trained model on an external independent CT dataset. The results show that our model significantly outperforms single-task alternatives, improving the mean surface distance from 7.174 mm to 4.196 mm. We also demonstrated that our approach is successful for different network architectures as backbones.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge