Liang Dai
Pangu Ultra MoE: How to Train Your Big MoE on Ascend NPUs
May 07, 2025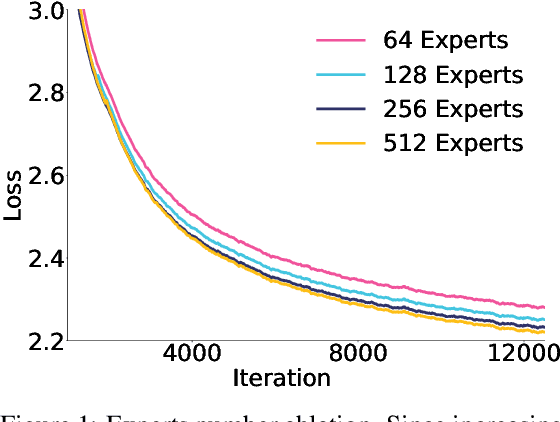

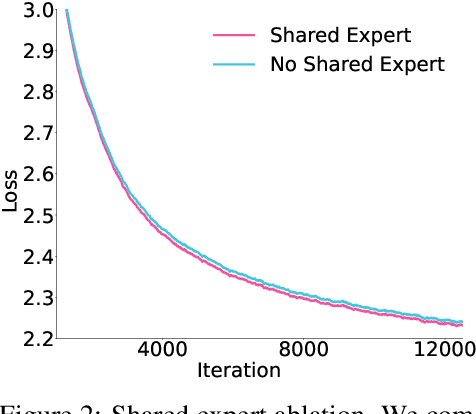

Abstract:Sparse large language models (LLMs) with Mixture of Experts (MoE) and close to a trillion parameters are dominating the realm of most capable language models. However, the massive model scale poses significant challenges for the underlying software and hardware systems. In this paper, we aim to uncover a recipe to harness such scale on Ascend NPUs. The key goals are better usage of the computing resources under the dynamic sparse model structures and materializing the expected performance gain on the actual hardware. To select model configurations suitable for Ascend NPUs without repeatedly running the expensive experiments, we leverage simulation to compare the trade-off of various model hyperparameters. This study led to Pangu Ultra MoE, a sparse LLM with 718 billion parameters, and we conducted experiments on the model to verify the simulation results. On the system side, we dig into Expert Parallelism to optimize the communication between NPU devices to reduce the synchronization overhead. We also optimize the memory efficiency within the devices to further reduce the parameter and activation management overhead. In the end, we achieve an MFU of 30.0% when training Pangu Ultra MoE, with performance comparable to that of DeepSeek R1, on 6K Ascend NPUs, and demonstrate that the Ascend system is capable of harnessing all the training stages of the state-of-the-art language models. Extensive experiments indicate that our recipe can lead to efficient training of large-scale sparse language models with MoE. We also study the behaviors of such models for future reference.
Rotatable Antenna-Enabled Secure Wireless Communication
Apr 15, 2025Abstract:Rotatable antenna (RA) is a promising technology that exploits new spatial degrees of freedom (DoFs) to improve wireless communication and sensing performance. In this letter, we investigate an RA-enabled secure communication system where confidential information is transmitted from an RA-based access point (AP) to a single-antenna legitimate user in the presence of multiple eavesdroppers. We aim to maximize the achievable secrecy rate by jointly optimizing the transmit beamforming and the deflection angles of all RAs. Accordingly, we propose an efficient alternating optimization (AO) algorithm to obtain a high-quality suboptimal solution in an iterative manner, where the generalized Rayleigh quotient-based beamforming is applied and the RAs' deflection angles are optimized by the successive convex approximation (SCA). Simulation results show that the proposed RA-enabled secure communication system achieves significant improvement in achievable secrecy rate as compared to various benchmark schemes.
Applying Deep Reinforcement Learning to the HP Model for Protein Structure Prediction
Dec 09, 2022



Abstract:A central problem in computational biophysics is protein structure prediction, i.e., finding the optimal folding of a given amino acid sequence. This problem has been studied in a classical abstract model, the HP model, where the protein is modeled as a sequence of H (hydrophobic) and P (polar) amino acids on a lattice. The objective is to find conformations maximizing H-H contacts. It is known that even in this reduced setting, the problem is intractable (NP-hard). In this work, we apply deep reinforcement learning (DRL) to the two-dimensional HP model. We can obtain the conformations of best known energies for benchmark HP sequences with lengths from 20 to 50. Our DRL is based on a deep Q-network (DQN). We find that a DQN based on long short-term memory (LSTM) architecture greatly enhances the RL learning ability and significantly improves the search process. DRL can sample the state space efficiently, without the need of manual heuristics. Experimentally we show that it can find multiple distinct best-known solutions per trial. This study demonstrates the effectiveness of deep reinforcement learning in the HP model for protein folding.
Sequential Monte Carlo Methods for System Identification
Mar 10, 2016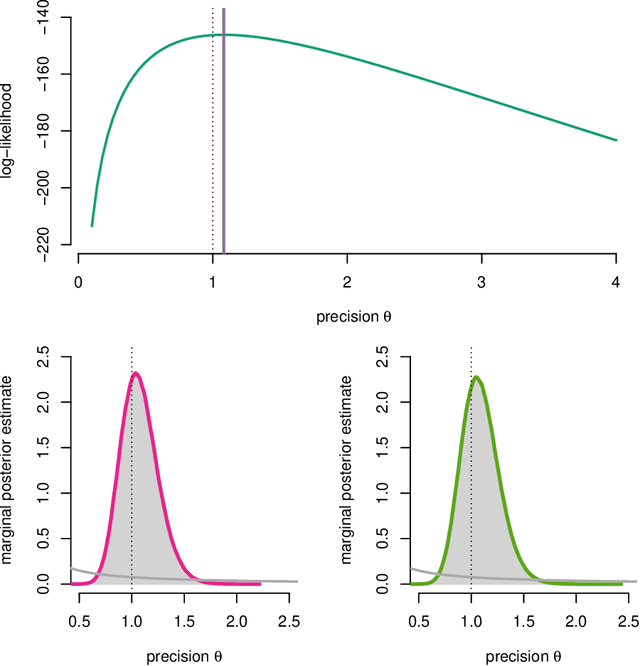
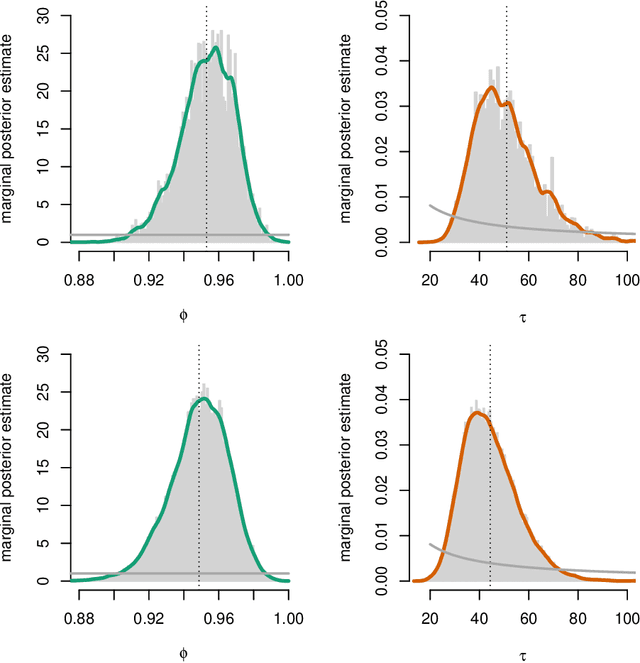
Abstract:One of the key challenges in identifying nonlinear and possibly non-Gaussian state space models (SSMs) is the intractability of estimating the system state. Sequential Monte Carlo (SMC) methods, such as the particle filter (introduced more than two decades ago), provide numerical solutions to the nonlinear state estimation problems arising in SSMs. When combined with additional identification techniques, these algorithms provide solid solutions to the nonlinear system identification problem. We describe two general strategies for creating such combinations and discuss why SMC is a natural tool for implementing these strategies.
Sparse Estimation From Noisy Observations of an Overdetermined Linear System
May 25, 2014



Abstract:This note studies a method for the efficient estimation of a finite number of unknown parameters from linear equations, which are perturbed by Gaussian noise. In case the unknown parameters have only few nonzero entries, the proposed estimator performs more efficiently than a traditional approach. The method consists of three steps: (1) a classical Least Squares Estimate (LSE), (2) the support is recovered through a Linear Programming (LP) optimization problem which can be computed using a soft-thresholding step, (3) a de-biasing step using a LSE on the estimated support set. The main contribution of this note is a formal derivation of an associated ORACLE property of the final estimate. That is, when the number of samples is large enough, the estimate is shown to equal the LSE based on the support of the {\em true} parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge