Automatic Foot Ulcer segmentation Using an Ensemble of Convolutional Neural Networks
Paper and Code
Sep 03, 2021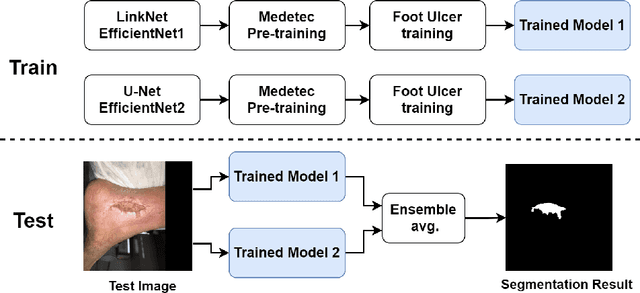
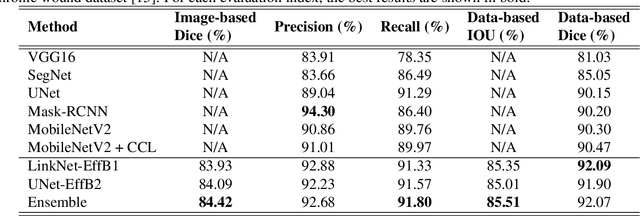
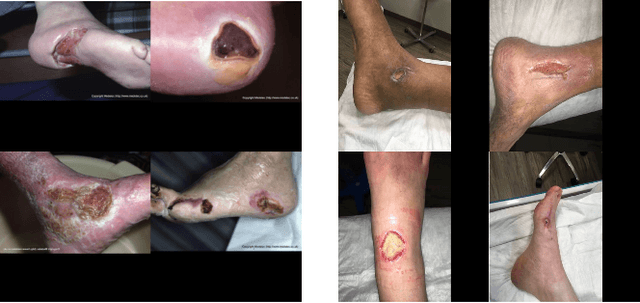

Foot ulcer is a common complication of diabetes mellitus; it is associated with substantial morbidity and mortality and remains a major risk factor for lower leg amputation. Extracting accurate morphological features from the foot wounds is crucial for proper treatment. Although visual and manual inspection by medical professionals is the common approach to extract the features, this method is subjective and error-prone. Computer-mediated approaches are the alternative solutions to segment the lesions and extract related morphological features. Among various proposed computer-based approaches for image segmentation, deep learning-based methods and more specifically convolutional neural networks (CNN) have shown excellent performances for various image segmentation tasks including medical image segmentation. In this work, we proposed an ensemble approach based on two encoder-decoder-based CNN models, namely LinkNet and UNet, to perform foot ulcer segmentation. To deal with limited training samples, we used pre-trained weights (EfficientNetB1 for the LinkNet model and EfficientNetB2 for the UNet model) and further pre-training by the Medetec dataset. We also applied a number of morphological-based and colour-based augmentation techniques to train the models. We integrated five-fold cross-validation, test time augmentation and result fusion in our proposed ensemble approach to boost the segmentation performance. Applied on a publicly available foot ulcer segmentation dataset and the MICCAI 2021 Foot Ulcer Segmentation (FUSeg) Challenge, our method achieved state-of-the-art data-based Dice scores of 92.07% and 88.80%, respectively. Our developed method achieved the first rank in the FUSeg challenge leaderboard. The Dockerised guideline, inference codes and saved trained models are publicly available in the published GitHub repository: https://github.com/masih4/Foot_Ulcer_Segmentation
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge