Vladimir Juras
Improved tissue sodium concentration quantification in breast cancer by reducing partial volume effects: a preliminary study
Mar 25, 2025



Abstract:Introduction: In sodium (23Na) MRI, partial volume effects (PVE) are one of the most common causes of errors in the quantification of tissue sodium concentration (TSC) in vivo. Advanced image reconstruction algorithms, such as compressed sensing (CS), have been shown to potentially reduce PVE. Therefore, we investigated the feasibility of CS-based methods for image quality and TSC quantification accuracy improvement in patients with breast cancer (BC). Subjects and Methods: Three healthy participants and 12 female participants with BC were examined on a 7T MRI scanner in this study. We reconstructed 23Na-MRI images using the weighted total variation (wTV) and directional total variation (dTV), anatomically guided total variation (AG-TV), and adaptive combine (ADC) reconstruction and performed image quality assessment. We evaluated agreement in tumor volumes delineated on sodium data using the Dice score and performed TSC quantification for different image reconstruction approaches. Results: All methods provided sodium images of the breast with good quality. The mean Dice scores for wTV, dTV, and AG-TV were 65%, 72%, and 75%, respectively. In the breast tumors, average TSC values were 83.0, 72.0, 80.0, and 84.0 mmol/L, respectively. There was a significant difference between dTV and wTV (p<0.001), as well as between dTV and AG-TV (p<0.001) and dTV and ADC algorithm (p<0.001). Conclusion: The results of this study showed that there are differences in tumor appearance and TSC estimations that might be depending on the type of image reconstruction and parameters used, most likely due to differences in their robustness in reducing PVE.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020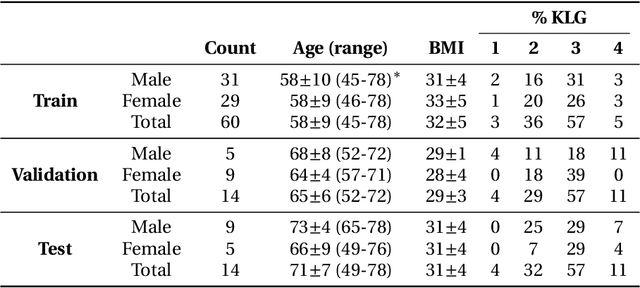
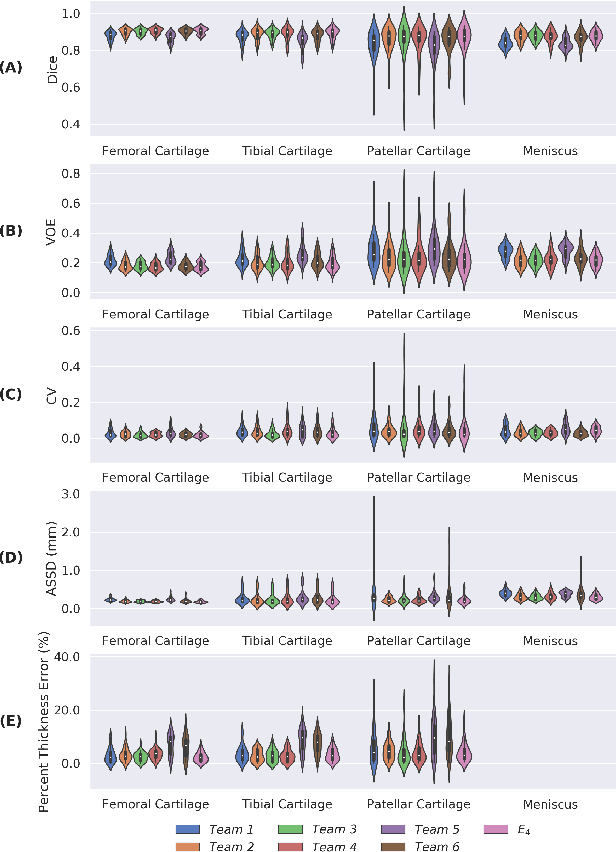
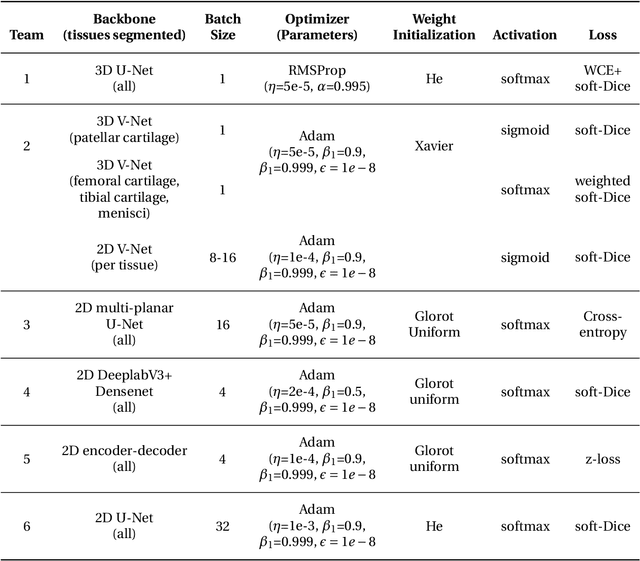
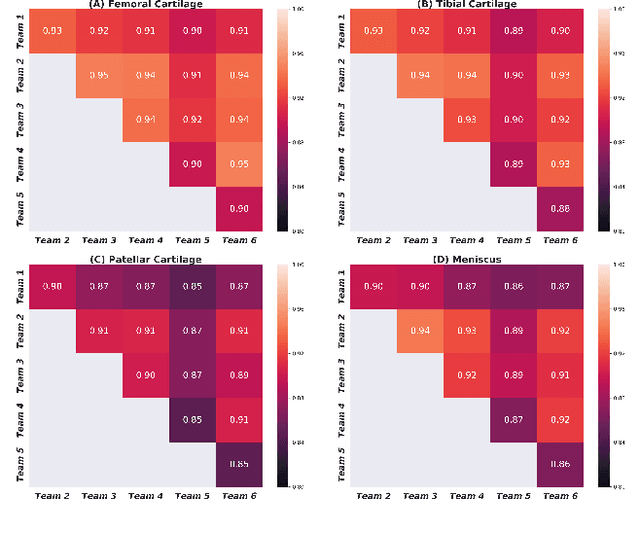
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge