Matthias Samwald
CSMeD: Bridging the Dataset Gap in Automated Citation Screening for Systematic Literature Reviews
Nov 21, 2023



Abstract:Systematic literature reviews (SLRs) play an essential role in summarising, synthesising and validating scientific evidence. In recent years, there has been a growing interest in using machine learning techniques to automate the identification of relevant studies for SLRs. However, the lack of standardised evaluation datasets makes comparing the performance of such automated literature screening systems difficult. In this paper, we analyse the citation screening evaluation datasets, revealing that many of the available datasets are either too small, suffer from data leakage or have limited applicability to systems treating automated literature screening as a classification task, as opposed to, for example, a retrieval or question-answering task. To address these challenges, we introduce CSMeD, a meta-dataset consolidating nine publicly released collections, providing unified access to 325 SLRs from the fields of medicine and computer science. CSMeD serves as a comprehensive resource for training and evaluating the performance of automated citation screening models. Additionally, we introduce CSMeD-FT, a new dataset designed explicitly for evaluating the full text publication screening task. To demonstrate the utility of CSMeD, we conduct experiments and establish baselines on new datasets.
An automatically discovered chain-of-thought prompt generalizes to novel models and datasets
May 04, 2023



Abstract:Emergent chain-of-thought (CoT) reasoning capabilities promise to improve performance and explainability of large language models (LLMs). However, uncertainties remain about how prompting strategies formulated for previous model generations generalize to new model generations and different datasets. In this small-scale study we compare the performance of a range of zero-shot prompts for inducing CoT reasoning across six recently released LLMs (davinci-002, davinci-003, GPT-3.5-turbo, GPT-4, Flan-T5-xxl and Cohere command-xlarge) on a mixture of six question-answering datasets, including datasets from scientific and medical domains. We find that a CoT prompt that was previously discovered through automated prompt discovery shows robust performance across experimental conditions and produces best results when applied to the state-of-the-art model GPT-4.
Model-agnostic explainable artificial intelligence for object detection in image data
Apr 12, 2023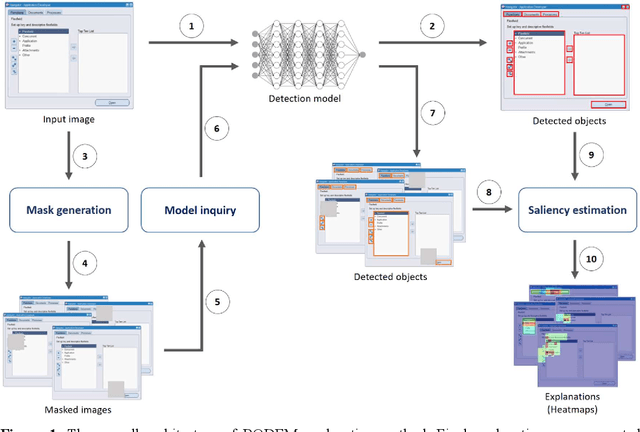
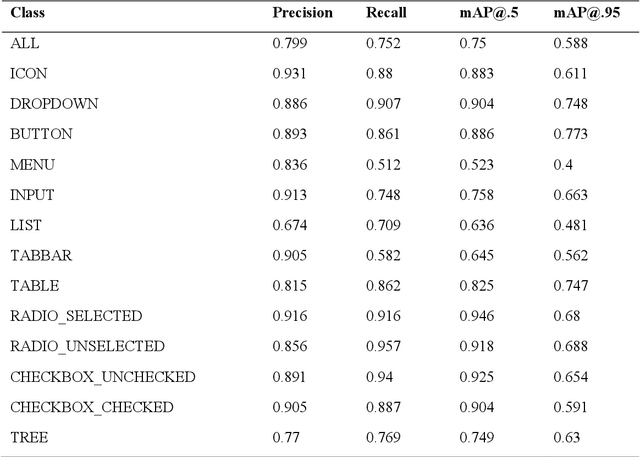
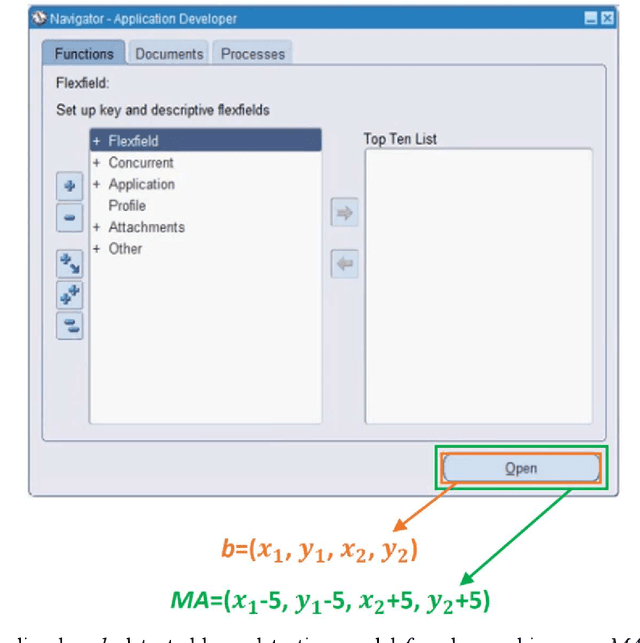
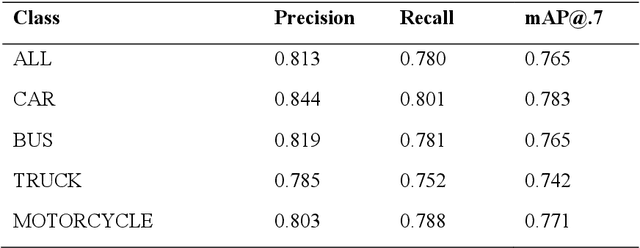
Abstract:Object detection is a fundamental task in computer vision, which has been greatly progressed through developing large and intricate deep learning models. However, the lack of transparency is a big challenge that may not allow the widespread adoption of these models. Explainable artificial intelligence is a field of research where methods are developed to help users understand the behavior, decision logics, and vulnerabilities of AI-based systems. Black-box explanation refers to explaining decisions of an AI system without having access to its internals. In this paper, we design and implement a black-box explanation method named Black-box Object Detection Explanation by Masking (BODEM) through adopting a new masking approach for AI-based object detection systems. We propose local and distant masking to generate multiple versions of an input image. Local masks are used to disturb pixels within a target object to figure out how the object detector reacts to these changes, while distant masks are used to assess how the detection model's decisions are affected by disturbing pixels outside the object. A saliency map is then created by estimating the importance of pixels through measuring the difference between the detection output before and after masking. Finally, a heatmap is created that visualizes how important pixels within the input image are to the detected objects. The experimentations on various object detection datasets and models showed that BODEM can be effectively used to explain the behavior of object detectors and reveal their vulnerabilities. This makes BODEM suitable for explaining and validating AI based object detection systems in black-box software testing scenarios. Furthermore, we conducted data augmentation experiments that showed local masks produced by BODEM can be used for further training the object detectors and improve their detection accuracy and robustness.
Applying unsupervised keyphrase methods on concepts extracted from discharge sheets
Mar 15, 2023Abstract:Clinical notes containing valuable patient information are written by different health care providers with various scientific levels and writing styles. It might be helpful for clinicians and researchers to understand what information is essential when dealing with extensive electronic medical records. Entities recognizing and mapping them to standard terminologies is crucial in reducing ambiguity in processing clinical notes. Although named entity recognition and entity linking are critical steps in clinical natural language processing, they can also result in the production of repetitive and low-value concepts. In other hand, all parts of a clinical text do not share the same importance or content in predicting the patient's condition. As a result, it is necessary to identify the section in which each content is recorded and also to identify key concepts to extract meaning from clinical texts. In this study, these challenges have been addressed by using clinical natural language processing techniques. In addition, in order to identify key concepts, a set of popular unsupervised key phrase extraction methods has been verified and evaluated. Considering that most of the clinical concepts are in the form of multi-word expressions and their accurate identification requires the user to specify n-gram range, we have proposed a shortcut method to preserve the structure of the expression based on TF-IDF. In order to evaluate the pre-processing method and select the concepts, we have designed two types of downstream tasks (multiple and binary classification) using the capabilities of transformer-based models. The obtained results show the superiority of proposed method in combination with SciBERT model, also offer an insight into the efficacy of general extracting essential phrase methods for clinical notes.
ThoughtSource: A central hub for large language model reasoning data
Jan 27, 2023Abstract:Large language models (LLMs) such as GPT-3 and ChatGPT have recently demonstrated impressive results across a wide range of tasks. LLMs are still limited, however, in that they frequently fail at complex reasoning, their reasoning processes are opaque, they are prone to 'hallucinate' facts, and there are concerns about their underlying biases. Letting models verbalize reasoning steps as natural language, a technique known as chain-of-thought prompting, has recently been proposed as a way to address some of these issues. Here we present the first release of ThoughtSource, a meta-dataset and software library for chain-of-thought (CoT) reasoning. The goal of ThoughtSource is to improve future artificial intelligence systems by facilitating qualitative understanding of CoTs, enabling empirical evaluations, and providing training data. This first release of ThoughtSource integrates six scientific/medical, three general-domain and five math word question answering datasets.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022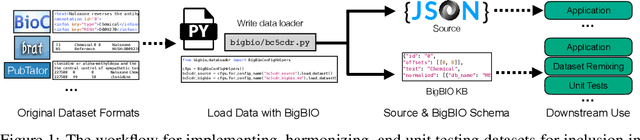
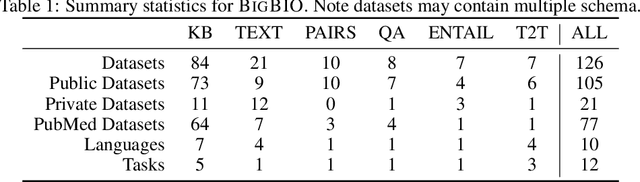
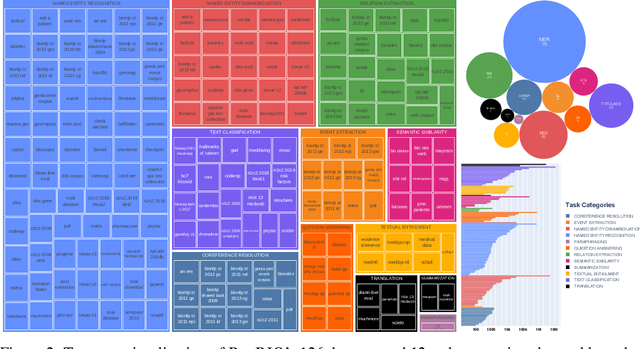
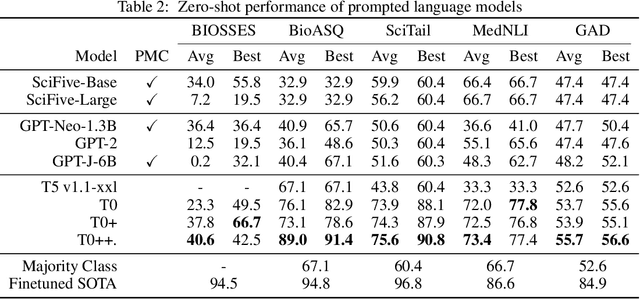
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
A global analysis of metrics used for measuring performance in natural language processing
Apr 25, 2022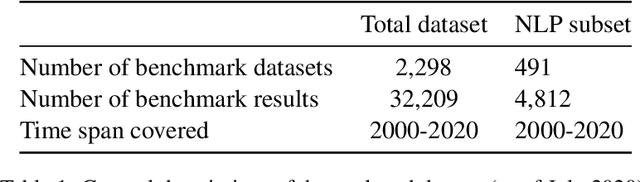
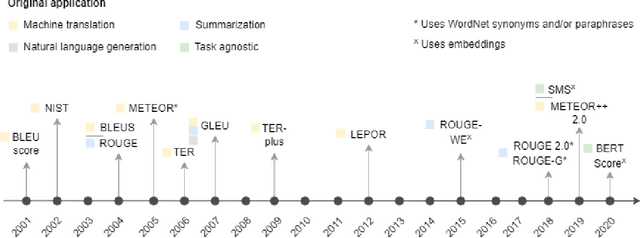
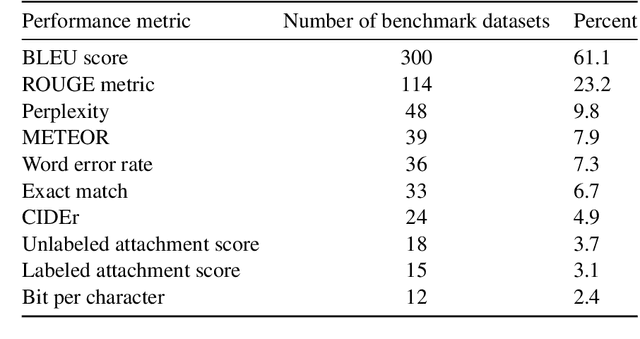
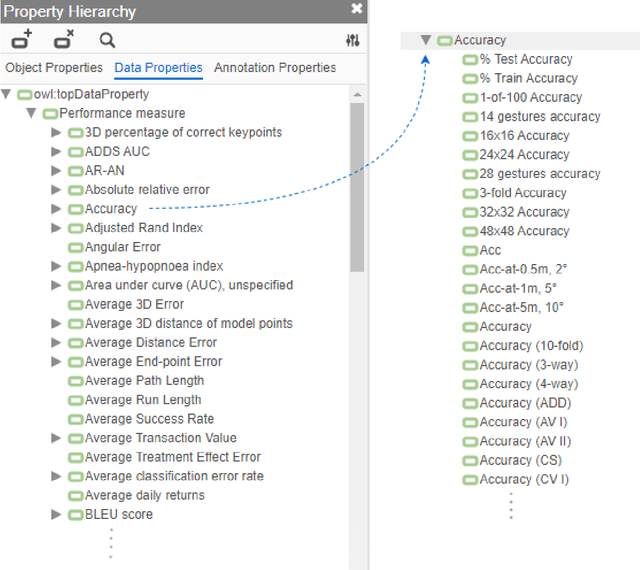
Abstract:Measuring the performance of natural language processing models is challenging. Traditionally used metrics, such as BLEU and ROUGE, originally devised for machine translation and summarization, have been shown to suffer from low correlation with human judgment and a lack of transferability to other tasks and languages. In the past 15 years, a wide range of alternative metrics have been proposed. However, it is unclear to what extent this has had an impact on NLP benchmarking efforts. Here we provide the first large-scale cross-sectional analysis of metrics used for measuring performance in natural language processing. We curated, mapped and systematized more than 3500 machine learning model performance results from the open repository 'Papers with Code' to enable a global and comprehensive analysis. Our results suggest that the large majority of natural language processing metrics currently used have properties that may result in an inadequate reflection of a models' performance. Furthermore, we found that ambiguities and inconsistencies in the reporting of metrics may lead to difficulties in interpreting and comparing model performances, impairing transparency and reproducibility in NLP research.
Mapping global dynamics of benchmark creation and saturation in artificial intelligence
Mar 09, 2022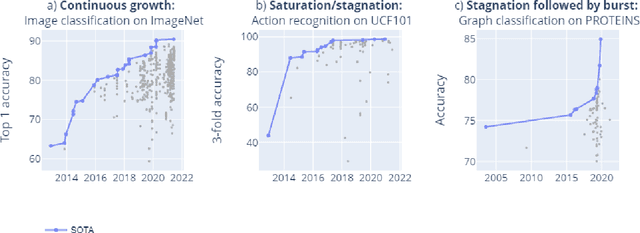
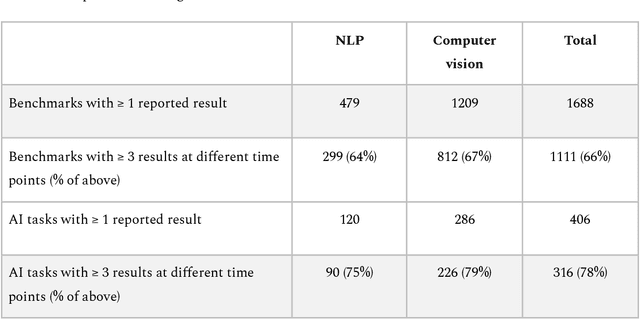
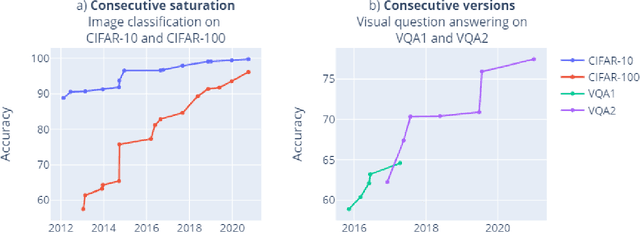
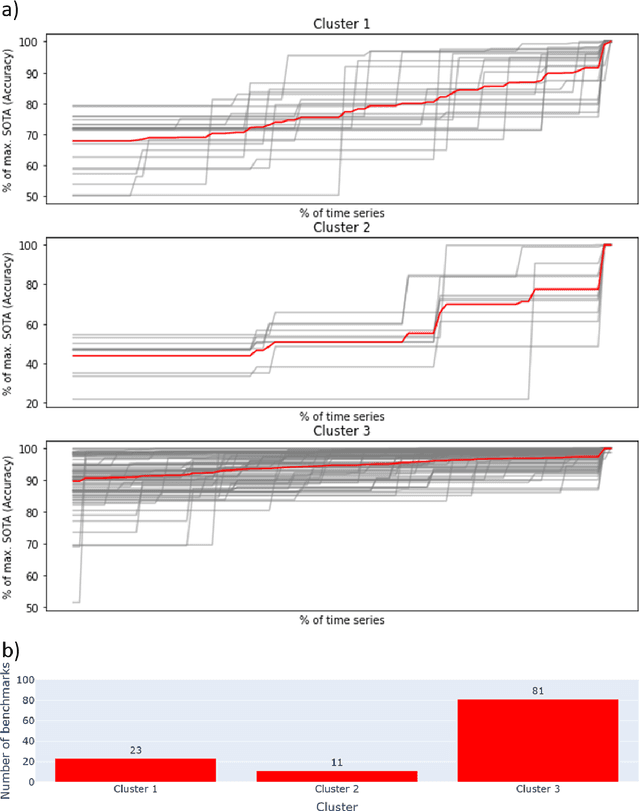
Abstract:Benchmarks are crucial to measuring and steering progress in artificial intelligence (AI). However, recent studies raised concerns over the state of AI benchmarking, reporting issues such as benchmark overfitting, benchmark saturation and increasing centralization of benchmark dataset creation. To facilitate monitoring of the health of the AI benchmarking ecosystem, we introduce methodologies for creating condensed maps of the global dynamics of benchmark creation and saturation. We curated data for 1688 benchmarks covering the entire domains of computer vision and natural language processing, and show that a large fraction of benchmarks quickly trended towards near-saturation, that many benchmarks fail to find widespread utilization, and that benchmark performance gains for different AI tasks were prone to unforeseen bursts. We conclude that future work should focus on large-scale community collaboration and on mapping benchmark performance gains to real-world utility and impact of AI.
Deep Learning, Natural Language Processing, and Explainable Artificial Intelligence in the Biomedical Domain
Mar 07, 2022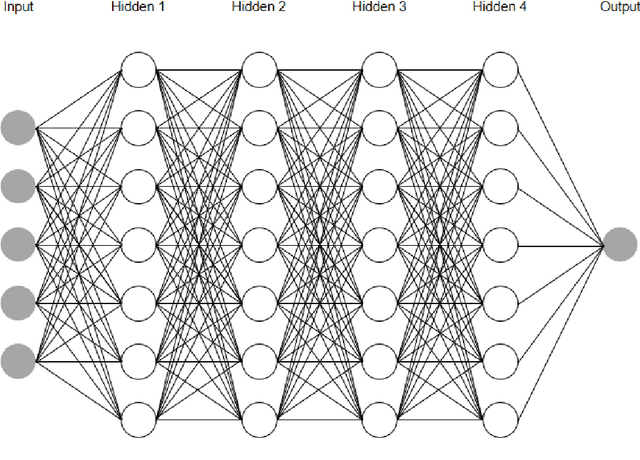
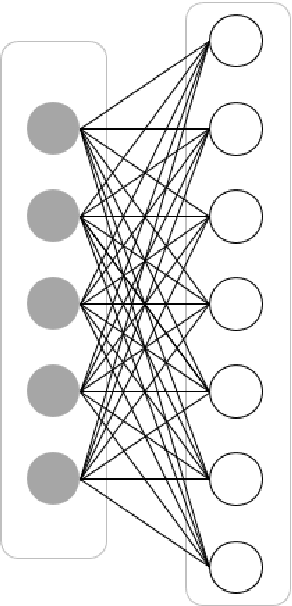
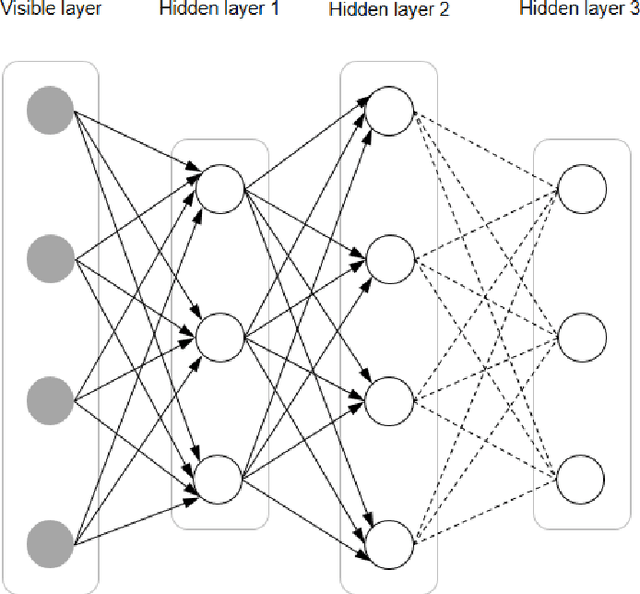
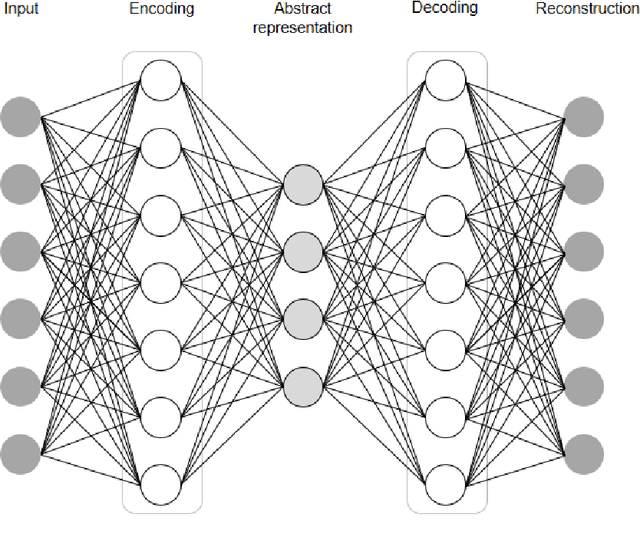
Abstract:In this article, we first give an introduction to artificial intelligence and its applications in biology and medicine in Section 1. Deep learning methods are then described in Section 2. We narrow down the focus of the study on textual data in Section 3, where natural language processing and its applications in the biomedical domain are described. In Section 4, we give an introduction to explainable artificial intelligence and discuss the importance of explainability of artificial intelligence systems, especially in the biomedical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge