Nasser Ghadiri
MedVQA-TREE: A Multimodal Reasoning and Retrieval Framework for Sarcopenia Prediction
Aug 26, 2025Abstract:Accurate sarcopenia diagnosis via ultrasound remains challenging due to subtle imaging cues, limited labeled data, and the absence of clinical context in most models. We propose MedVQA-TREE, a multimodal framework that integrates a hierarchical image interpretation module, a gated feature-level fusion mechanism, and a novel multi-hop, multi-query retrieval strategy. The vision module includes anatomical classification, region segmentation, and graph-based spatial reasoning to capture coarse, mid-level, and fine-grained structures. A gated fusion mechanism selectively integrates visual features with textual queries, while clinical knowledge is retrieved through a UMLS-guided pipeline accessing PubMed and a sarcopenia-specific external knowledge base. MedVQA-TREE was trained and evaluated on two public MedVQA datasets (VQA-RAD and PathVQA) and a custom sarcopenia ultrasound dataset. The model achieved up to 99% diagnostic accuracy and outperformed previous state-of-the-art methods by over 10%. These results underscore the benefit of combining structured visual understanding with guided knowledge retrieval for effective AI-assisted diagnosis in sarcopenia.
Data Optimisation of Machine Learning Models for Smart Irrigation in Urban Parks
Oct 03, 2024Abstract:Urban environments face significant challenges due to climate change, including extreme heat, drought, and water scarcity, which impact public health, community well-being, and local economies. Effective management of these issues is crucial, particularly in areas like Sydney Olympic Park, which relies on one of Australia's largest irrigation systems. The Smart Irrigation Management for Parks and Cool Towns (SIMPaCT) project, initiated in 2021, leverages advanced technologies and machine learning models to optimize irrigation and induce physical cooling. This paper introduces two novel methods to enhance the efficiency of the SIMPaCT system's extensive sensor network and applied machine learning models. The first method employs clustering of sensor time series data using K-shape and K-means algorithms to estimate readings from missing sensors, ensuring continuous and reliable data. This approach can detect anomalies, correct data sources, and identify and remove redundant sensors to reduce maintenance costs. The second method involves sequential data collection from different sensor locations using robotic systems, significantly reducing the need for high numbers of stationary sensors. Together, these methods aim to maintain accurate soil moisture predictions while optimizing sensor deployment and reducing maintenance costs, thereby enhancing the efficiency and effectiveness of the smart irrigation system. Our evaluations demonstrate significant improvements in the efficiency and cost-effectiveness of soil moisture monitoring networks. The cluster-based replacement of missing sensors provides up to 5.4% decrease in average error. The sequential sensor data collection as a robotic emulation shows 17.2% and 2.1% decrease in average error for circular and linear paths respectively.
PerkwE_COQA: Enhanced Persian Conversational Question Answering by combining contextual keyword extraction with Large Language Models
Apr 15, 2024Abstract:Smart cities need the involvement of their residents to enhance quality of life. Conversational query-answering is an emerging approach for user engagement. There is an increasing demand of an advanced conversational question-answering that goes beyond classic systems. Existing approaches have shown that LLMs offer promising capabilities for CQA, but may struggle to capture the nuances of conversational contexts. The new approach involves understanding the content and engaging in a multi-step conversation with the user to fulfill their needs. This paper presents a novel method to elevate the performance of Persian Conversational question-answering (CQA) systems. It combines the strengths of Large Language Models (LLMs) with contextual keyword extraction. Our method extracts keywords specific to the conversational flow, providing the LLM with additional context to understand the user's intent and generate more relevant and coherent responses. We evaluated the effectiveness of this combined approach through various metrics, demonstrating significant improvements in CQA performance compared to an LLM-only baseline. The proposed method effectively handles implicit questions, delivers contextually relevant answers, and tackles complex questions that rely heavily on conversational context. The findings indicate that our method outperformed the evaluation benchmarks up to 8% higher than existing methods and the LLM-only baseline.
Multi-level biomedical NER through multi-granularity embeddings and enhanced labeling
Dec 24, 2023Abstract:Biomedical Named Entity Recognition (NER) is a fundamental task of Biomedical Natural Language Processing for extracting relevant information from biomedical texts, such as clinical records, scientific publications, and electronic health records. The conventional approaches for biomedical NER mainly use traditional machine learning techniques, such as Conditional Random Fields and Support Vector Machines or deep learning-based models like Recurrent Neural Networks and Convolutional Neural Networks. Recently, Transformer-based models, including BERT, have been used in the domain of biomedical NER and have demonstrated remarkable results. However, these models are often based on word-level embeddings, limiting their ability to capture character-level information, which is effective in biomedical NER due to the high variability and complexity of biomedical texts. To address these limitations, this paper proposes a hybrid approach that integrates the strengths of multiple models. In this paper, we proposed an approach that leverages fine-tuned BERT to provide contextualized word embeddings, a pre-trained multi-channel CNN for character-level information capture, and following by a BiLSTM + CRF for sequence labelling and modelling dependencies between the words in the text. In addition, also we propose an enhanced labelling method as part of pre-processing to enhance the identification of the entity's beginning word and thus improve the identification of multi-word entities, a common challenge in biomedical NER. By integrating these models and the pre-processing method, our proposed model effectively captures both contextual information and detailed character-level information. We evaluated our model on the benchmark i2b2/2010 dataset, achieving an F1-score of 90.11. These results illustrate the proficiency of our proposed model in performing biomedical Named Entity Recognition.
Applying unsupervised keyphrase methods on concepts extracted from discharge sheets
Mar 15, 2023Abstract:Clinical notes containing valuable patient information are written by different health care providers with various scientific levels and writing styles. It might be helpful for clinicians and researchers to understand what information is essential when dealing with extensive electronic medical records. Entities recognizing and mapping them to standard terminologies is crucial in reducing ambiguity in processing clinical notes. Although named entity recognition and entity linking are critical steps in clinical natural language processing, they can also result in the production of repetitive and low-value concepts. In other hand, all parts of a clinical text do not share the same importance or content in predicting the patient's condition. As a result, it is necessary to identify the section in which each content is recorded and also to identify key concepts to extract meaning from clinical texts. In this study, these challenges have been addressed by using clinical natural language processing techniques. In addition, in order to identify key concepts, a set of popular unsupervised key phrase extraction methods has been verified and evaluated. Considering that most of the clinical concepts are in the form of multi-word expressions and their accurate identification requires the user to specify n-gram range, we have proposed a shortcut method to preserve the structure of the expression based on TF-IDF. In order to evaluate the pre-processing method and select the concepts, we have designed two types of downstream tasks (multiple and binary classification) using the capabilities of transformer-based models. The obtained results show the superiority of proposed method in combination with SciBERT model, also offer an insight into the efficacy of general extracting essential phrase methods for clinical notes.
Assessing mortality prediction through different representation models based on concepts extracted from clinical notes
Jul 22, 2022



Abstract:Recent years have seen particular interest in using electronic medical records (EMRs) for secondary purposes to enhance the quality and safety of healthcare delivery. EMRs tend to contain large amounts of valuable clinical notes. Learning of embedding is a method for converting notes into a format that makes them comparable. Transformer-based representation models have recently made a great leap forward. These models are pre-trained on large online datasets to understand natural language texts effectively. The quality of a learning embedding is influenced by how clinical notes are used as input to representation models. A clinical note has several sections with different levels of information value. It is also common for healthcare providers to use different expressions for the same concept. Existing methods use clinical notes directly or with an initial preprocessing as input to representation models. However, to learn a good embedding, we identified the most essential clinical notes section. We then mapped the extracted concepts from selected sections to the standard names in the Unified Medical Language System (UMLS). We used the standard phrases corresponding to the unique concepts as input for clinical models. We performed experiments to measure the usefulness of the learned embedding vectors in the task of hospital mortality prediction on a subset of the publicly available Medical Information Mart for Intensive Care (MIMIC-III) dataset. According to the experiments, clinical transformer-based representation models produced better results with getting input generated by standard names of extracted unique concepts compared to other input formats. The best-performing models were BioBERT, PubMedBERT, and UmlsBERT, respectively.
Integration of Text and Graph-based Features for Detecting Mental Health Disorders from Voice
May 14, 2022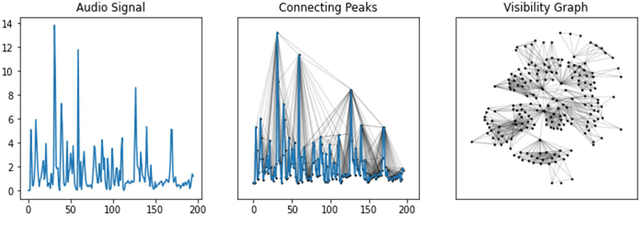
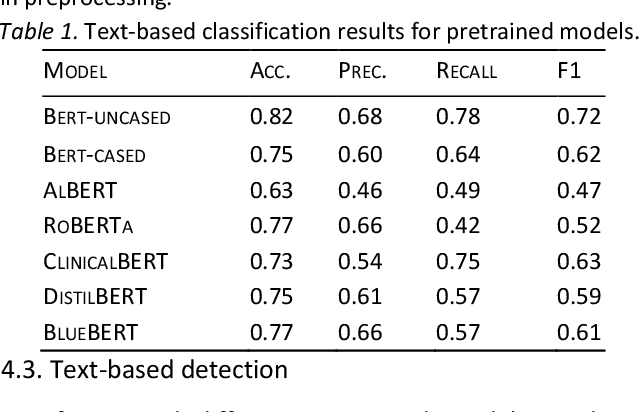
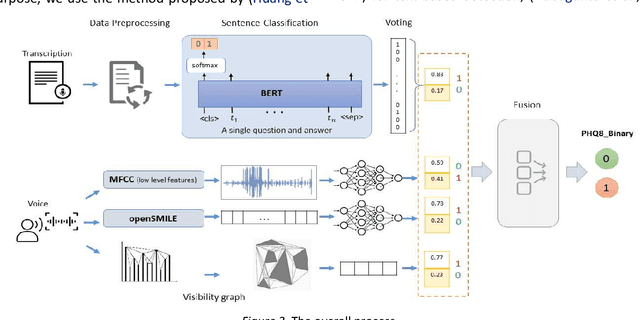
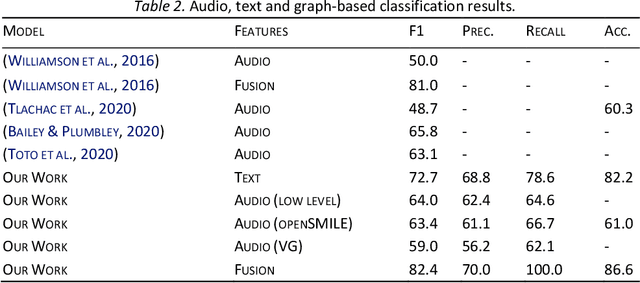
Abstract:With the availability of voice-enabled devices such as smart phones, mental health disorders could be detected and treated earlier, particularly post-pandemic. The current methods involve extracting features directly from audio signals. In this paper, two methods are used to enrich voice analysis for depression detection: graph transformation of voice signals, and natural language processing of the transcript based on representational learning, fused together to produce final class labels. The results of experiments with the DAIC-WOZ dataset suggest that integration of text-based voice classification and learning from low level and graph-based voice signal features can improve the detection of mental disorders like depression.
A complex network approach to time series analysis with application in diagnosis of neuromuscular disorders
Aug 16, 2021
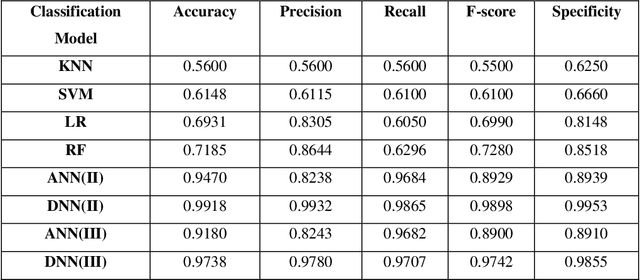


Abstract:Electromyography (EMG) refers to a biomedical signal indicating neuromuscular activity and muscle morphology. Experts accurately diagnose neuromuscular disorders using this time series. Modern data analysis techniques have recently led to introducing novel approaches for mapping time series data to graphs and complex networks with applications in diverse fields, including medicine. The resulting networks develop a completely different visual acuity that can be used to complement physician findings of time series. This can lead to a more enriched analysis, reduced error, more accurate diagnosis of the disease, and increased accuracy and speed of the treatment process. The mapping process may cause the loss of essential data from the time series and not retain all the time series features. As a result, achieving an approach that can provide a good representation of the time series while maintaining essential features is crucial. This paper proposes a new approach to network development named GraphTS to overcome the limited accuracy of existing methods through EMG time series using the visibility graph method. For this purpose, EMG signals are pre-processed and mapped to a complex network by a standard visibility graph algorithm. The resulting networks can differentiate between healthy and patient samples. In the next step, the properties of the developed networks are given in the form of a feature matrix as input to classifiers after extracting optimal features. Performance evaluation of the proposed approach with deep neural network shows 99.30% accuracy for training data and 99.18% for test data. Therefore, in addition to enriched network representation and covering the features of time series for healthy, myopathy, and neuropathy EMG, the proposed technique improves accuracy, precision, recall, and F-score.
Hybrid deep learning methods for phenotype prediction from clinical notes
Aug 16, 2021
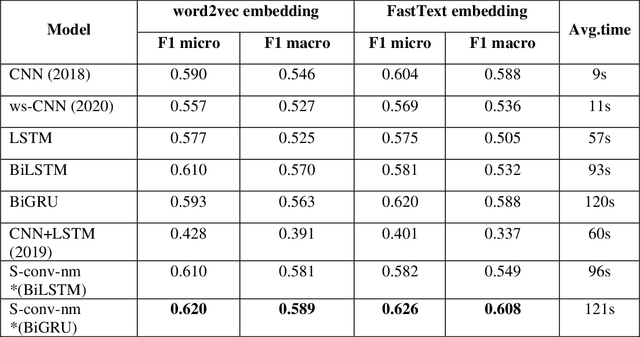

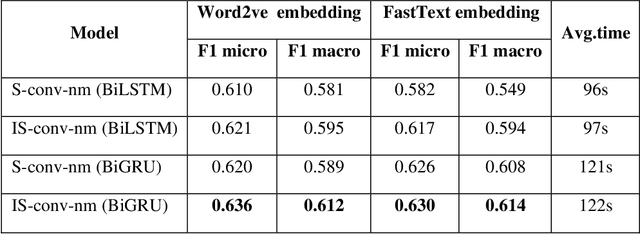
Abstract:Identifying patient cohorts from clinical notes in secondary electronic health records is a fundamental task in clinical information management. The patient cohort identification needs to identify the patient phenotypes. However, with the growing number of clinical notes, it becomes challenging to analyze the data manually. Therefore, automatic extraction of clinical concepts would be an essential task to identify the patient phenotypes correctly. This paper proposes a novel hybrid model for automatically extracting patient phenotypes using natural language processing and deep learning models to determine the patient phenotypes without dictionaries and human intervention. The proposed hybrid model is based on a neural bidirectional sequence model (BiLSTM or BiGRU) and a Convolutional Neural Network (CNN) for identifying patient's phenotypes in discharge reports. Furthermore, to extract more features related to each phenotype, an extra CNN layer is run parallel to the hybrid proposed model. We used pre-trained embeddings such as FastText and Word2vec separately as the input layers to evaluate other embedding's performance in identifying patient phenotypes. We also measured the effect of applying additional data cleaning steps on discharge reports to identify patient phenotypes by deep learning models. We used discharge reports in the Medical Information Mart for Intensive Care III (MIMIC III) database. Experimental results in internal comparison demonstrate significant performance improvement over existing models. The enhanced model with an extra CNN layer obtained a relatively higher F1-score than the original hybrid model.
Heterogeneous electronic medical record representation for similarity computing
Apr 29, 2021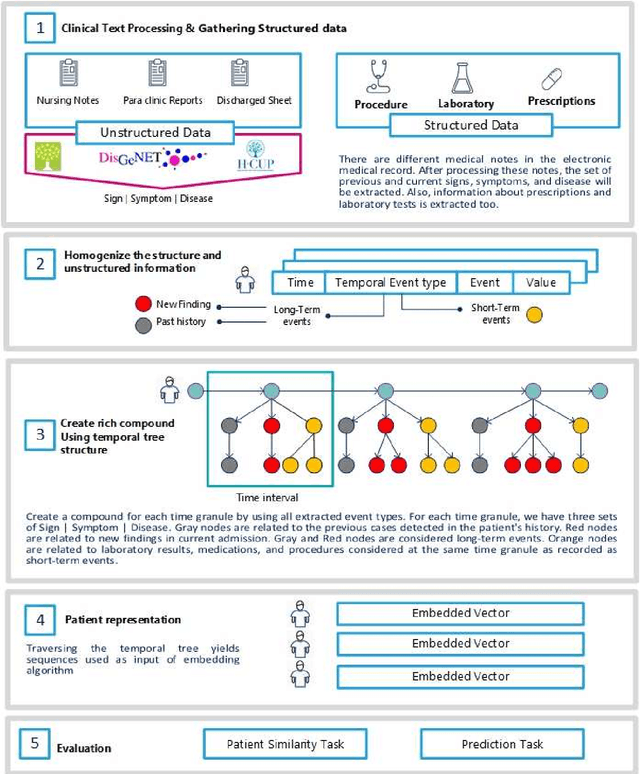
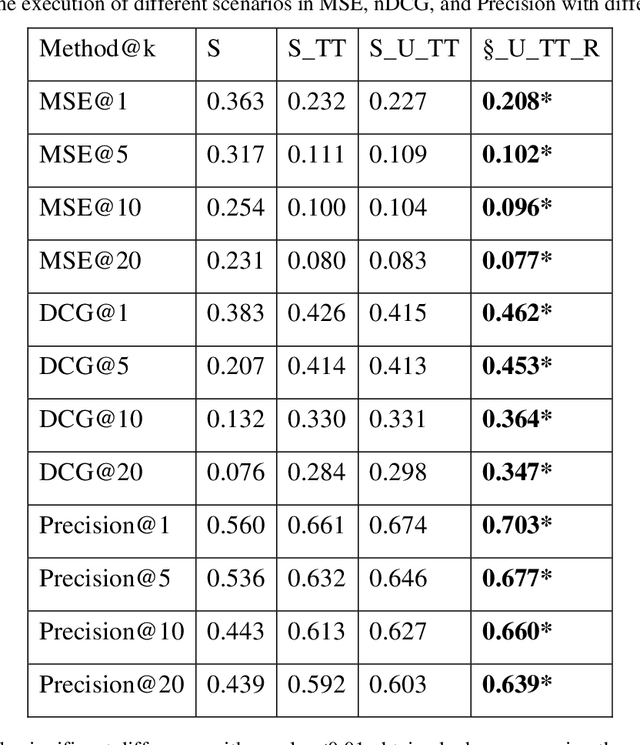


Abstract:Due to the widespread use of tools and the development of text processing techniques, the size and range of clinical data are not limited to structured data. The rapid growth of recorded information has led to big data platforms in healthcare that could be used to improve patients' primary care and serve various secondary purposes. Patient similarity assessment is one of the secondary tasks in identifying patients who are similar to a given patient, and it helps derive insights from similar patients' records to provide better treatment. This type of assessment is based on calculating the distance between patients. Since representing and calculating the similarity of patients plays an essential role in many secondary uses of electronic records, this article examines a new data representation method for Electronic Medical Records (EMRs) while taking into account the information in clinical narratives for similarity computing. Some previous works are based on structured data types, while other works only use unstructured data. However, a comprehensive representation of the information contained in the EMR requires the effective aggregation of both structured and unstructured data. To address the limitations of previous methods, we propose a method that captures the co-occurrence of different medical events, including signs, symptoms, and diseases extracted via unstructured data and structured data. It integrates data as discriminative features to construct a temporal tree, considering the difference between events that have short-term and long-term impacts. Our results show that considering signs, symptoms, and diseases in every time interval leads to less MSE and more precision compared to baseline representations that do not consider this information or consider them separately from structured data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge