Sahar Khalafi
Designing an Improved Deep Learning-based Model for COVID-19 Recognition in Chest X-ray Images: A Knowledge Distillation Approach
Jan 06, 2023



Abstract:COVID-19 has adversely affected humans and societies in different aspects. Numerous people have perished due to inaccurate COVID-19 identification and, consequently, a lack of appropriate medical treatment. Numerous solutions based on manual and automatic feature extraction techniques have been investigated to address this issue by researchers worldwide. Typically, automatic feature extraction methods, particularly deep learning models, necessitate a powerful hardware system to perform the necessary computations. Unfortunately, many institutions and societies cannot benefit from these advancements due to the prohibitively high cost of high-quality hardware equipment. As a result, this study focused on two primary goals: first, lowering the computational costs associated with running the proposed model on embedded devices, mobile devices, and conventional computers; and second, improving the model's performance in comparison to previously published methods (at least performs on par with state-of-the-art models) in order to ensure its performance and accuracy for the medical recognition task. This study used two neural networks to improve feature extraction from our dataset: VGG19 and ResNet50V2. Both of these networks are capable of providing semantic features from the nominated dataset. To this end, An alternative network was considered, namely MobileNetV2, which excels at extracting semantic features while requiring minimal computation on mobile and embedded devices. Knowledge distillation (KD) was used to transfer knowledge from the teacher network (concatenated ResNet50V2 and VGG19) to the student network (MobileNetV2) to improve MobileNetV2 performance and to achieve a robust and accurate model for the COVID-19 identification task from chest X-ray images.
Hybrid deep learning methods for phenotype prediction from clinical notes
Aug 16, 2021
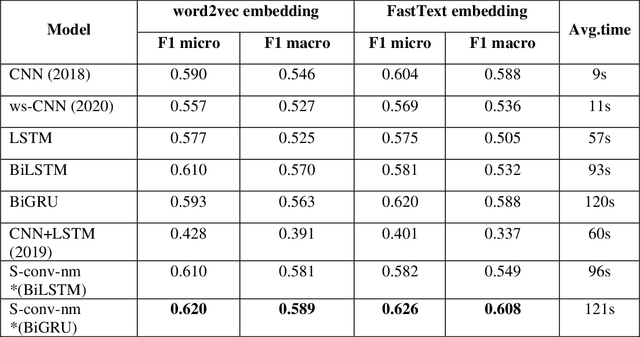

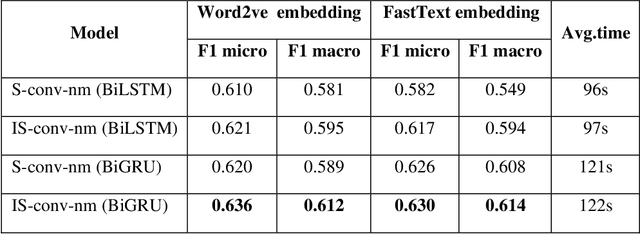
Abstract:Identifying patient cohorts from clinical notes in secondary electronic health records is a fundamental task in clinical information management. The patient cohort identification needs to identify the patient phenotypes. However, with the growing number of clinical notes, it becomes challenging to analyze the data manually. Therefore, automatic extraction of clinical concepts would be an essential task to identify the patient phenotypes correctly. This paper proposes a novel hybrid model for automatically extracting patient phenotypes using natural language processing and deep learning models to determine the patient phenotypes without dictionaries and human intervention. The proposed hybrid model is based on a neural bidirectional sequence model (BiLSTM or BiGRU) and a Convolutional Neural Network (CNN) for identifying patient's phenotypes in discharge reports. Furthermore, to extract more features related to each phenotype, an extra CNN layer is run parallel to the hybrid proposed model. We used pre-trained embeddings such as FastText and Word2vec separately as the input layers to evaluate other embedding's performance in identifying patient phenotypes. We also measured the effect of applying additional data cleaning steps on discharge reports to identify patient phenotypes by deep learning models. We used discharge reports in the Medical Information Mart for Intensive Care III (MIMIC III) database. Experimental results in internal comparison demonstrate significant performance improvement over existing models. The enhanced model with an extra CNN layer obtained a relatively higher F1-score than the original hybrid model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge