Fahime Shahrokh
Prediction of readmission of patients by extracting biomedical concepts from clinical texts
Mar 12, 2024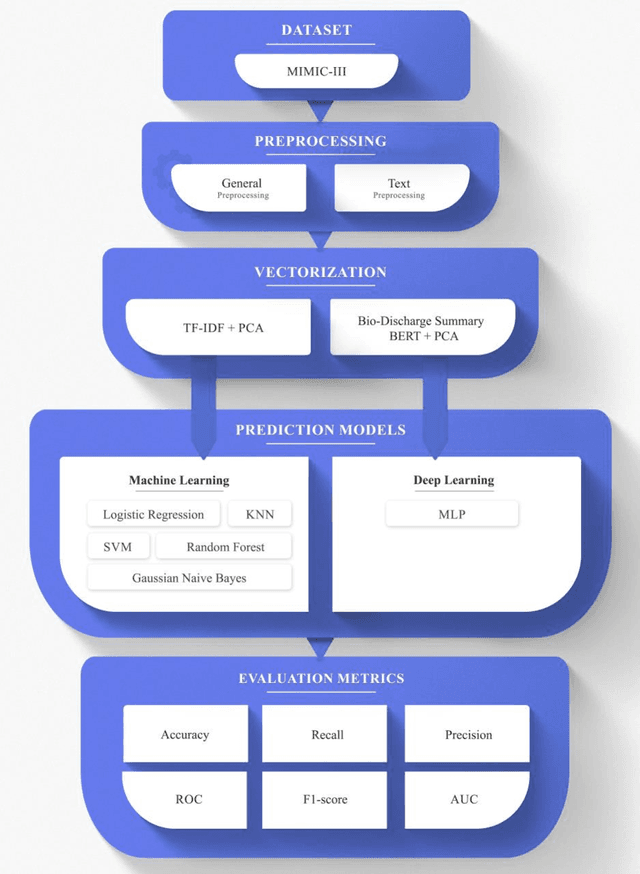


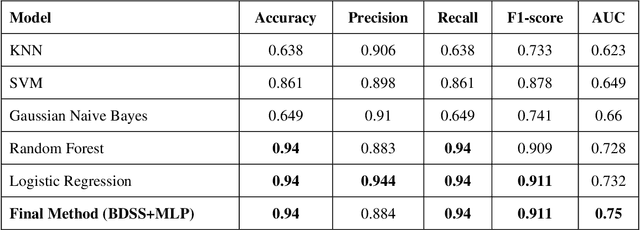
Abstract:Today, the existence of a vast amount of electronic health data has created potential capacities for conducting studies aiming to improve the medical services provided to patients and reduce the costs of the healthcare system. One of the topics that has been receiving attention in the field of medicine in recent years is the identification of patients who are likely to be re-hospitalized shortly after being discharged from the hospital. This identification can help doctors choose appropriate treatment methods, thereby reducing the rate of patient re-hospitalization and resulting in effective treatment cost reduction. In this study, the prediction of patient re-hospitalization using text mining approaches and the processing of discharge report texts in the patient's electronic file has been discussed. To this end, the performance of various machine learning models has been evaluated using two approaches: bag of word and bag of concept, in the process of predicting patient readmission. Comparing the efficiency of these approaches has shown the superiority of the random forest model and the bag of concept approach over other machine learning models and approaches. This research has achieved the highest score in predicting the probability of patient re-hospitalization, with a recall score of 68.9%, compared to similar works that have utilized machine learning models in this field.
Multi-level biomedical NER through multi-granularity embeddings and enhanced labeling
Dec 24, 2023Abstract:Biomedical Named Entity Recognition (NER) is a fundamental task of Biomedical Natural Language Processing for extracting relevant information from biomedical texts, such as clinical records, scientific publications, and electronic health records. The conventional approaches for biomedical NER mainly use traditional machine learning techniques, such as Conditional Random Fields and Support Vector Machines or deep learning-based models like Recurrent Neural Networks and Convolutional Neural Networks. Recently, Transformer-based models, including BERT, have been used in the domain of biomedical NER and have demonstrated remarkable results. However, these models are often based on word-level embeddings, limiting their ability to capture character-level information, which is effective in biomedical NER due to the high variability and complexity of biomedical texts. To address these limitations, this paper proposes a hybrid approach that integrates the strengths of multiple models. In this paper, we proposed an approach that leverages fine-tuned BERT to provide contextualized word embeddings, a pre-trained multi-channel CNN for character-level information capture, and following by a BiLSTM + CRF for sequence labelling and modelling dependencies between the words in the text. In addition, also we propose an enhanced labelling method as part of pre-processing to enhance the identification of the entity's beginning word and thus improve the identification of multi-word entities, a common challenge in biomedical NER. By integrating these models and the pre-processing method, our proposed model effectively captures both contextual information and detailed character-level information. We evaluated our model on the benchmark i2b2/2010 dataset, achieving an F1-score of 90.11. These results illustrate the proficiency of our proposed model in performing biomedical Named Entity Recognition.
Integration of Text and Graph-based Features for Detecting Mental Health Disorders from Voice
May 14, 2022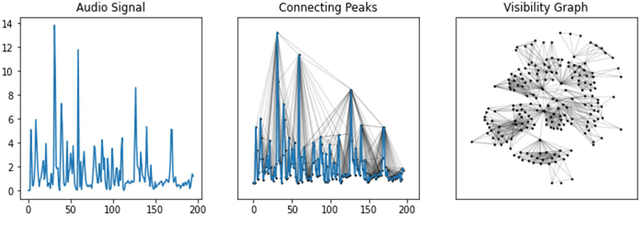
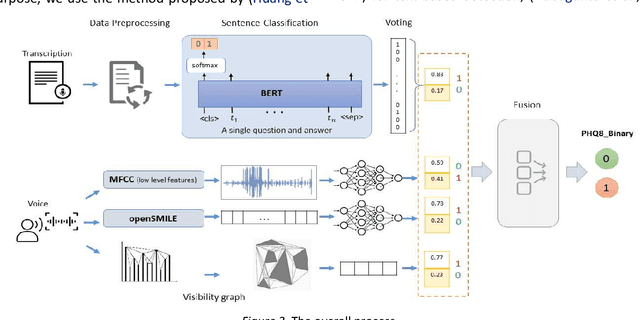
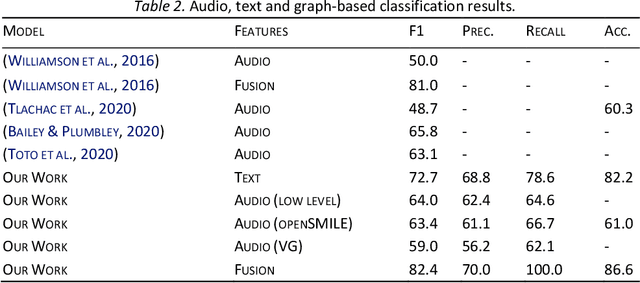
Abstract:With the availability of voice-enabled devices such as smart phones, mental health disorders could be detected and treated earlier, particularly post-pandemic. The current methods involve extracting features directly from audio signals. In this paper, two methods are used to enrich voice analysis for depression detection: graph transformation of voice signals, and natural language processing of the transcript based on representational learning, fused together to produce final class labels. The results of experiments with the DAIC-WOZ dataset suggest that integration of text-based voice classification and learning from low level and graph-based voice signal features can improve the detection of mental disorders like depression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge