Fabio Rinaldi
University of Zurich, Switzerland, Dalle Molle Institute for Artificial Intelligence Research, Swiss Institute of Bioinformatics, Switzerland
Are the LLMs Capable of Maintaining at Least the Language Genus?
Oct 24, 2025Abstract:Large Language Models (LLMs) display notable variation in multilingual behavior, yet the role of genealogical language structure in shaping this variation remains underexplored. In this paper, we investigate whether LLMs exhibit sensitivity to linguistic genera by extending prior analyses on the MultiQ dataset. We first check if models prefer to switch to genealogically related languages when prompt language fidelity is not maintained. Next, we investigate whether knowledge consistency is better preserved within than across genera. We show that genus-level effects are present but strongly conditioned by training resource availability. We further observe distinct multilingual strategies across LLMs families. Our findings suggest that LLMs encode aspects of genus-level structure, but training data imbalances remain the primary factor shaping their multilingual performance.
Boosting Radiology Report Generation by Infusing Comparison Prior
May 08, 2023



Abstract:Current transformer-based models achieved great success in generating radiology reports from chest X-ray images. Nonetheless, one of the major issues is the model's lack of prior knowledge, which frequently leads to false references to non-existent prior exams in synthetic reports. This is mainly due to the knowledge gap between radiologists and the generation models: radiologists are aware of the prior information of patients to write a medical report, while models only receive X-ray images at a specific time. To address this issue, we propose a novel approach that employs a labeler to extract comparison prior information from radiology reports in the IU X-ray and MIMIC-CXR datasets. This comparison prior is then incorporated into state-of-the-art transformer-based models, allowing them to generate more realistic and comprehensive reports. We test our method on the IU X-ray and MIMIC-CXR datasets and find that it outperforms previous state-of-the-art models in terms of both automatic and human evaluation metrics. In addition, unlike previous models, our model generates reports that do not contain false references to non-existent prior exams. Our approach provides a promising direction for bridging the gap between radiologists and generation models in medical report generation.
SST-BERT at SemEval-2020 Task 1: Semantic Shift Tracing by Clustering in BERT-based Embedding Spaces
Oct 02, 2020

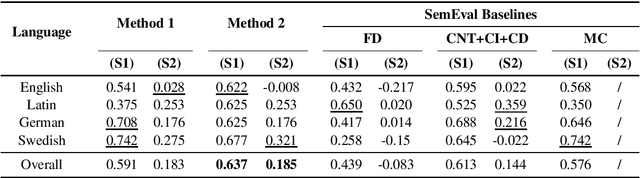
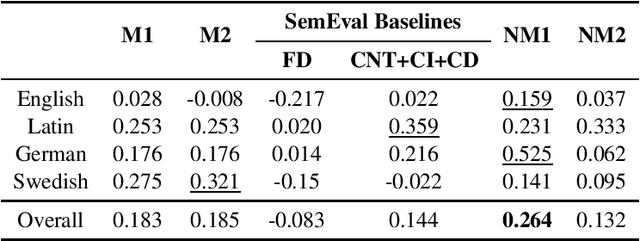
Abstract:Lexical semantic change detection (also known as semantic shift tracing) is a task of identifying words that have changed their meaning over time. Unsupervised semantic shift tracing, focal point of SemEval2020, is particularly challenging. Given the unsupervised setup, in this work, we propose to identify clusters among different occurrences of each target word, considering these as representatives of different word meanings. As such, disagreements in obtained clusters naturally allow to quantify the level of semantic shift per each target word in four target languages. To leverage this idea, clustering is performed on contextualized (BERT-based) embeddings of word occurrences. The obtained results show that our approach performs well both measured separately (per language) and overall, where we surpass all provided SemEval baselines.
MultiGBS: A multi-layer graph approach to biomedical summarization
Aug 27, 2020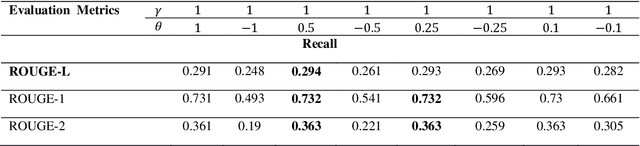
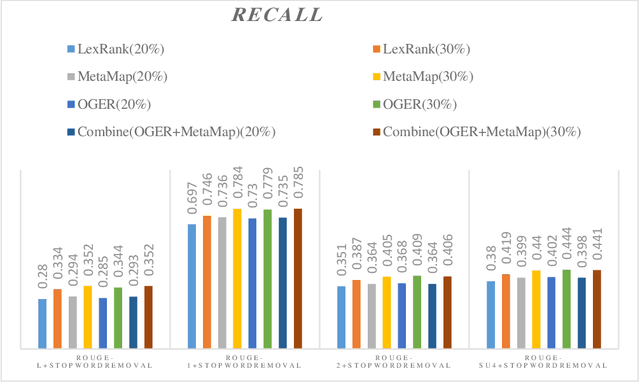

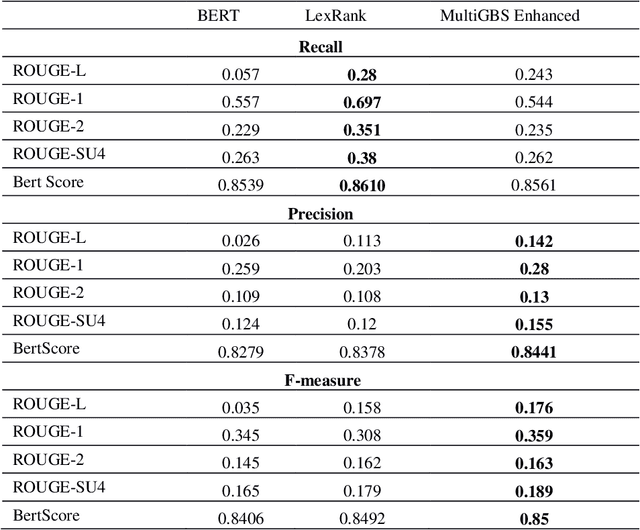
Abstract:Automatic text summarization methods generate a shorter version of the input text to assist the reader in gaining a quick yet informative gist. Existing text summarization methods generally focus on a single aspect of text when selecting the sentences, causing potential loss of essential information. We propose a domain-specific method that models a document as a multi-layer graph to enable processing multiple features of the text at the same time. The features we used in this paper are word similarity, semantic similarity, and co-reference similarity that are modeled as three different layers. The summarizer selects the sentences from the multi-layer graph based on the MultiRank algorithm and length of concepts. The proposed MultiGBS algorithm employs UMLS and extracts concepts and relationships with different tools such as SemRep, MetaMap, and OGER. Extensive evaluation by ROUGE and BertScore shows increased F-measure values. Compared with leveraging BERT as extractive text summarization, the improvements in F-measure are 0.141 for ROUGE-L, 0.014 for ROUGE-1, 0.018 for ROUGE-2, 0.024 for ROUGE-SU4, and 0.0094 for BertScore.
Parallel sequence tagging for concept recognition
Mar 16, 2020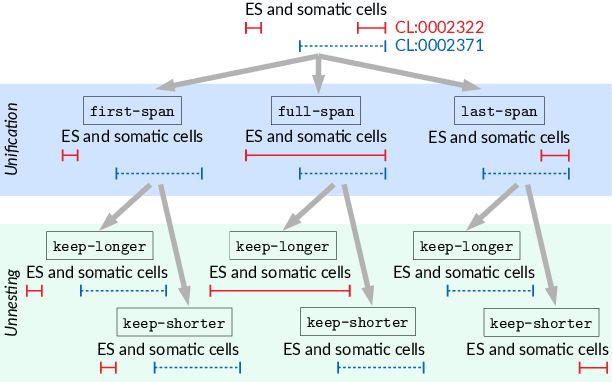
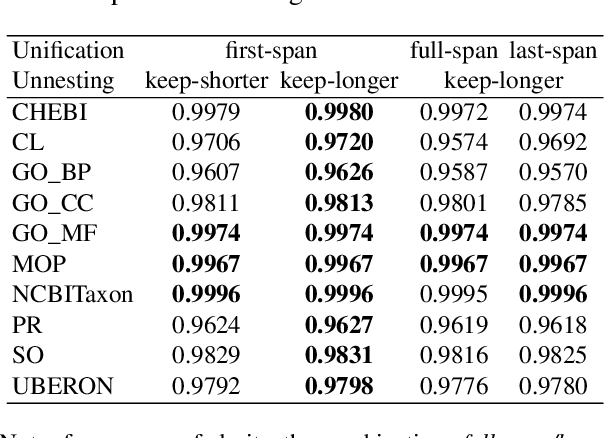
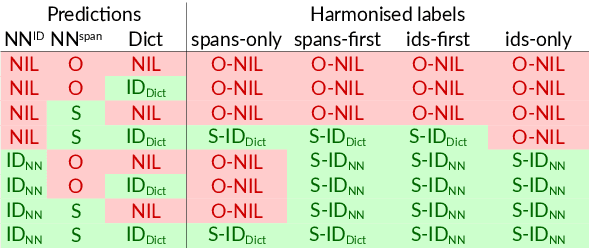
Abstract:Motivation: Named Entity Recognition (NER) and Normalisation (NEN) are core components of any text-mining system for biomedical texts. In a traditional concept-recognition pipeline, these tasks are combined in a serial way, which is inherently prone to error propagation from NER to NEN. We propose a parallel architecture, where both NER and NEN are modeled as a sequence-labeling task, operating directly on the source text. We examine different harmonisation strategies for merging the predictions of the two classifiers into a single output sequence. Results: We test our approach on the recent Version 4 of the CRAFT corpus. In all 20 annotation sets of the concept-annotation task, our system outperforms the pipeline system reported as a baseline in the CRAFT shared task 2019. Our analysis shows that the strengths of the two classifiers can be combined in a fruitful way. However, prediction harmonisation requires individual calibration on a development set for each annotation set. This allows achieving a good trade-off between established knowledge (training set) and novel information (unseen concepts). Availability and Implementation: Source code freely available for download at https://github.com/OntoGene/craft-st. Supplementary data are available at arXiv online.
Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review
Aug 15, 2019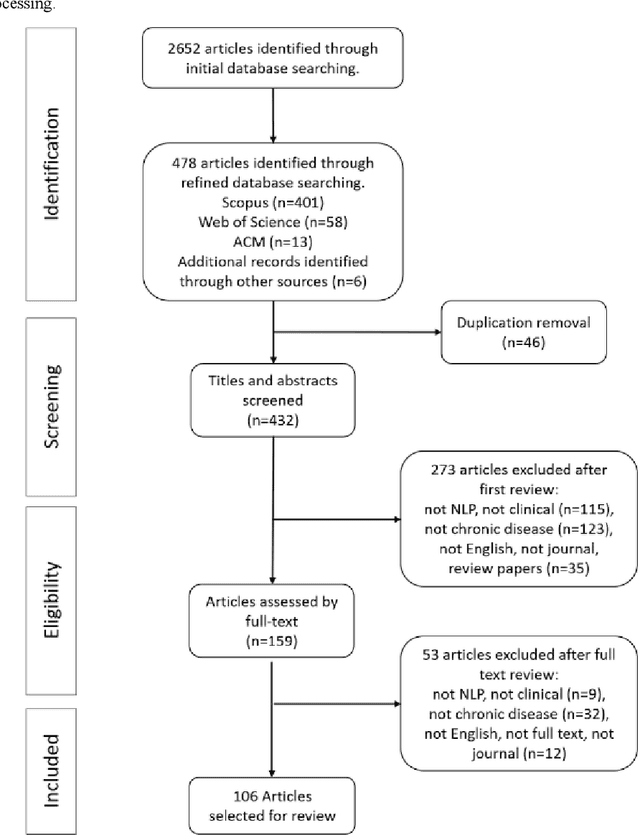

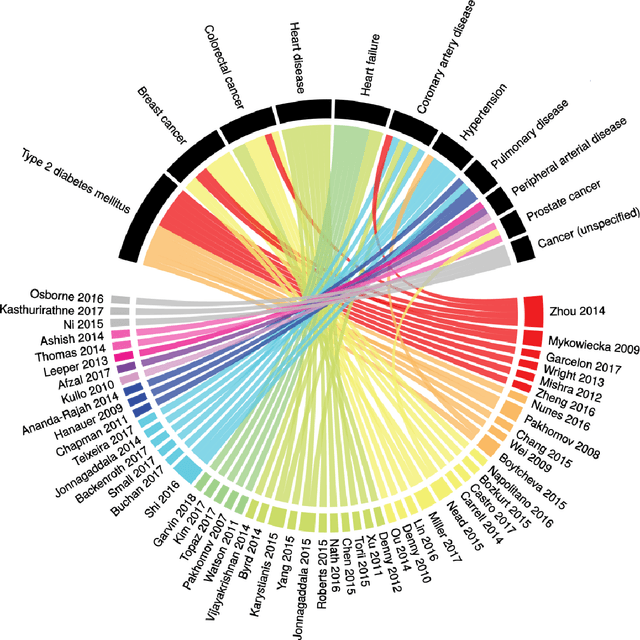

Abstract:Of the 2652 articles considered, 106 met the inclusion criteria. Review of the included papers resulted in identification of 43 chronic diseases, which were then further classified into 10 disease categories using ICD-10. The majority of studies focused on diseases of the circulatory system (n=38) while endocrine and metabolic diseases were fewest (n=14). This was due to the structure of clinical records related to metabolic diseases, which typically contain much more structured data, compared with medical records for diseases of the circulatory system, which focus more on unstructured data and consequently have seen a stronger focus of NLP. The review has shown that there is a significant increase in the use of machine learning methods compared to rule-based approaches; however, deep learning methods remain emergent (n=3). Consequently, the majority of works focus on classification of disease phenotype with only a handful of papers addressing extraction of comorbidities from the free text or integration of clinical notes with structured data. There is a notable use of relatively simple methods, such as shallow classifiers (or combination with rule-based methods), due to the interpretability of predictions, which still represents a significant issue for more complex methods. Finally, scarcity of publicly available data may also have contributed to insufficient development of more advanced methods, such as extraction of word embeddings from clinical notes. Further efforts are still required to improve (1) progression of clinical NLP methods from extraction toward understanding; (2) recognition of relations among entities rather than entities in isolation; (3) temporal extraction to understand past, current, and future clinical events; (4) exploitation of alternative sources of clinical knowledge; and (5) availability of large-scale, de-identified clinical corpora.
* Supplementary material detailing articles reviewed, classification of diseases and associated algorithms, can be found at: http://venetosmani.com/research/publications.html
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge