Seyedmostafa Sheikhalishahi
Benchmarking machine learning models on eICU critical care dataset
Oct 02, 2019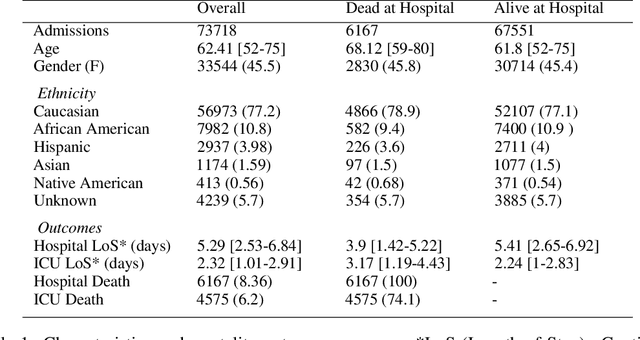
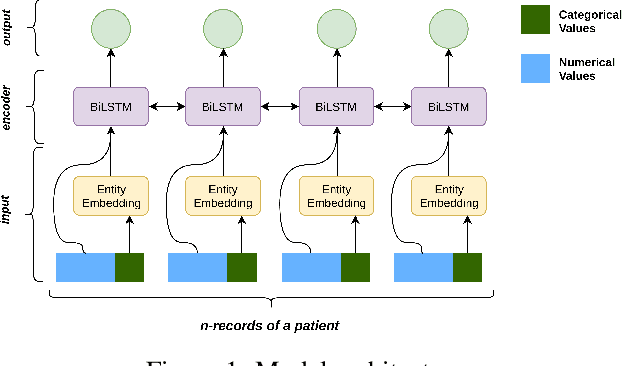

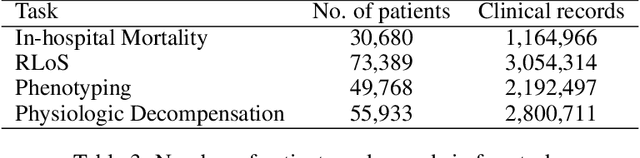
Abstract:Progress of machine learning in critical care has been difficult to track, in part due to absence of public benchmarks. Other fields of research (such as vision and NLP) have already established various competitions and benchmarks, whereas only recent availability of large clinical datasets has enabled the possibility of public benchmarks. Taking advantage of this opportunity, we propose a public benchmark suite to address four areas of critical care, namely mortality prediction, estimation of length of stay, patient phenotyping and risk of decompensation. We define each task and compare the performance of both clinical models as well as baseline and deep models using eICU critical care dataset of around 73,000 patients. Furthermore, we investigate the impact of numerical variables as well as handling of categorical variables for each of the defined tasks.
Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review
Aug 15, 2019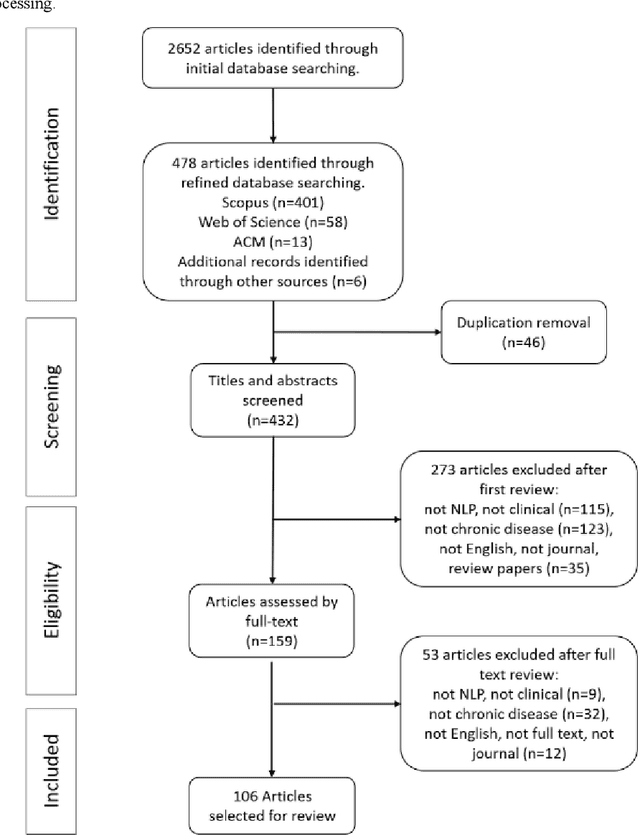

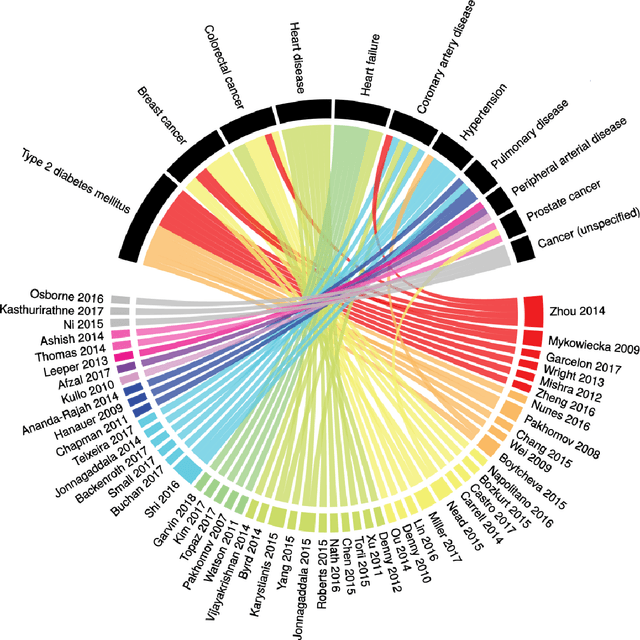

Abstract:Of the 2652 articles considered, 106 met the inclusion criteria. Review of the included papers resulted in identification of 43 chronic diseases, which were then further classified into 10 disease categories using ICD-10. The majority of studies focused on diseases of the circulatory system (n=38) while endocrine and metabolic diseases were fewest (n=14). This was due to the structure of clinical records related to metabolic diseases, which typically contain much more structured data, compared with medical records for diseases of the circulatory system, which focus more on unstructured data and consequently have seen a stronger focus of NLP. The review has shown that there is a significant increase in the use of machine learning methods compared to rule-based approaches; however, deep learning methods remain emergent (n=3). Consequently, the majority of works focus on classification of disease phenotype with only a handful of papers addressing extraction of comorbidities from the free text or integration of clinical notes with structured data. There is a notable use of relatively simple methods, such as shallow classifiers (or combination with rule-based methods), due to the interpretability of predictions, which still represents a significant issue for more complex methods. Finally, scarcity of publicly available data may also have contributed to insufficient development of more advanced methods, such as extraction of word embeddings from clinical notes. Further efforts are still required to improve (1) progression of clinical NLP methods from extraction toward understanding; (2) recognition of relations among entities rather than entities in isolation; (3) temporal extraction to understand past, current, and future clinical events; (4) exploitation of alternative sources of clinical knowledge; and (5) availability of large-scale, de-identified clinical corpora.
* Supplementary material detailing articles reviewed, classification of diseases and associated algorithms, can be found at: http://venetosmani.com/research/publications.html
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge