Mizuho Nishio
Department of Radiology, Kobe University Graduate School of Medicine, Kobe, Japan
RadVLM: A Multitask Conversational Vision-Language Model for Radiology
Feb 05, 2025



Abstract:The widespread use of chest X-rays (CXRs), coupled with a shortage of radiologists, has driven growing interest in automated CXR analysis and AI-assisted reporting. While existing vision-language models (VLMs) show promise in specific tasks such as report generation or abnormality detection, they often lack support for interactive diagnostic capabilities. In this work we present RadVLM, a compact, multitask conversational foundation model designed for CXR interpretation. To this end, we curate a large-scale instruction dataset comprising over 1 million image-instruction pairs containing both single-turn tasks -- such as report generation, abnormality classification, and visual grounding -- and multi-turn, multi-task conversational interactions. After fine-tuning RadVLM on this instruction dataset, we evaluate it across different tasks along with re-implemented baseline VLMs. Our results show that RadVLM achieves state-of-the-art performance in conversational capabilities and visual grounding while remaining competitive in other radiology tasks. Ablation studies further highlight the benefit of joint training across multiple tasks, particularly for scenarios with limited annotated data. Together, these findings highlight the potential of RadVLM as a clinically relevant AI assistant, providing structured CXR interpretation and conversational capabilities to support more effective and accessible diagnostic workflows.
Exploring Multilingual Large Language Models for Enhanced TNM classification of Radiology Report in lung cancer staging
Jun 12, 2024Abstract:Background: Structured radiology reports remains underdeveloped due to labor-intensive structuring and narrative-style reporting. Deep learning, particularly large language models (LLMs) like GPT-3.5, offers promise in automating the structuring of radiology reports in natural languages. However, although it has been reported that LLMs are less effective in languages other than English, their radiological performance has not been extensively studied. Purpose: This study aimed to investigate the accuracy of TNM classification based on radiology reports using GPT3.5-turbo (GPT3.5) and the utility of multilingual LLMs in both Japanese and English. Material and Methods: Utilizing GPT3.5, we developed a system to automatically generate TNM classifications from chest CT reports for lung cancer and evaluate its performance. We statistically analyzed the impact of providing full or partial TNM definitions in both languages using a Generalized Linear Mixed Model. Results: Highest accuracy was attained with full TNM definitions and radiology reports in English (M = 94%, N = 80%, T = 47%, and ALL = 36%). Providing definitions for each of the T, N, and M factors statistically improved their respective accuracies (T: odds ratio (OR) = 2.35, p < 0.001; N: OR = 1.94, p < 0.01; M: OR = 2.50, p < 0.001). Japanese reports exhibited decreased N and M accuracies (N accuracy: OR = 0.74 and M accuracy: OR = 0.21). Conclusion: This study underscores the potential of multilingual LLMs for automatic TNM classification in radiology reports. Even without additional model training, performance improvements were evident with the provided TNM definitions, indicating LLMs' relevance in radiology contexts.
Radiology-Aware Model-Based Evaluation Metric for Report Generation
Nov 28, 2023Abstract:We propose a new automated evaluation metric for machine-generated radiology reports using the successful COMET architecture adapted for the radiology domain. We train and publish four medically-oriented model checkpoints, including one trained on RadGraph, a radiology knowledge graph. Our results show that our metric correlates moderately to high with established metrics such as BERTscore, BLEU, and CheXbert scores. Furthermore, we demonstrate that one of our checkpoints exhibits a high correlation with human judgment, as assessed using the publicly available annotations of six board-certified radiologists, using a set of 200 reports. We also performed our own analysis gathering annotations with two radiologists on a collection of 100 reports. The results indicate the potential effectiveness of our method as a radiology-specific evaluation metric. The code, data, and model checkpoints to reproduce our findings will be publicly available.
Development of pericardial fat count images using a combination of three different deep-learning models
Jul 25, 2023



Abstract:Rationale and Objectives: Pericardial fat (PF), the thoracic visceral fat surrounding the heart, promotes the development of coronary artery disease by inducing inflammation of the coronary arteries. For evaluating PF, this study aimed to generate pericardial fat count images (PFCIs) from chest radiographs (CXRs) using a dedicated deep-learning model. Materials and Methods: The data of 269 consecutive patients who underwent coronary computed tomography (CT) were reviewed. Patients with metal implants, pleural effusion, history of thoracic surgery, or that of malignancy were excluded. Thus, the data of 191 patients were used. PFCIs were generated from the projection of three-dimensional CT images, where fat accumulation was represented by a high pixel value. Three different deep-learning models, including CycleGAN, were combined in the proposed method to generate PFCIs from CXRs. A single CycleGAN-based model was used to generate PFCIs from CXRs for comparison with the proposed method. To evaluate the image quality of the generated PFCIs, structural similarity index measure (SSIM), mean squared error (MSE), and mean absolute error (MAE) of (i) the PFCI generated using the proposed method and (ii) the PFCI generated using the single model were compared. Results: The mean SSIM, MSE, and MAE were as follows: 0.856, 0.0128, and 0.0357, respectively, for the proposed model; and 0.762, 0.0198, and 0.0504, respectively, for the single CycleGAN-based model. Conclusion: PFCIs generated from CXRs with the proposed model showed better performance than those with the single model. PFCI evaluation without CT may be possible with the proposed method.
Boosting Radiology Report Generation by Infusing Comparison Prior
May 08, 2023



Abstract:Current transformer-based models achieved great success in generating radiology reports from chest X-ray images. Nonetheless, one of the major issues is the model's lack of prior knowledge, which frequently leads to false references to non-existent prior exams in synthetic reports. This is mainly due to the knowledge gap between radiologists and the generation models: radiologists are aware of the prior information of patients to write a medical report, while models only receive X-ray images at a specific time. To address this issue, we propose a novel approach that employs a labeler to extract comparison prior information from radiology reports in the IU X-ray and MIMIC-CXR datasets. This comparison prior is then incorporated into state-of-the-art transformer-based models, allowing them to generate more realistic and comprehensive reports. We test our method on the IU X-ray and MIMIC-CXR datasets and find that it outperforms previous state-of-the-art models in terms of both automatic and human evaluation metrics. In addition, unlike previous models, our model generates reports that do not contain false references to non-existent prior exams. Our approach provides a promising direction for bridging the gap between radiologists and generation models in medical report generation.
Unsupervised-learning-based method for chest MRI-CT transformation using structure constrained unsupervised generative attention networks
Jun 16, 2021



Abstract:The integrated positron emission tomography/magnetic resonance imaging (PET/MRI) scanner facilitates the simultaneous acquisition of metabolic information via PET and morphological information with high soft-tissue contrast using MRI. Although PET/MRI facilitates the capture of high-accuracy fusion images, its major drawback can be attributed to the difficulty encountered when performing attenuation correction, which is necessary for quantitative PET evaluation. The combined PET/MRI scanning requires the generation of attenuation-correction maps from MRI owing to no direct relationship between the gamma-ray attenuation information and MRIs. While MRI-based bone-tissue segmentation can be readily performed for the head and pelvis regions, the realization of accurate bone segmentation via chest CT generation remains a challenging task. This can be attributed to the respiratory and cardiac motions occurring in the chest as well as its anatomically complicated structure and relatively thin bone cortex. This paper presents a means to minimise the anatomical structural changes without human annotation by adding structural constraints using a modality-independent neighbourhood descriptor (MIND) to a generative adversarial network (GAN) that can transform unpaired images. The results obtained in this study revealed the proposed U-GAT-IT + MIND approach to outperform all other competing approaches. The findings of this study hint towards possibility of synthesising clinically acceptable CT images from chest MRI without human annotation, thereby minimising the changes in the anatomical structure.
Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray image: combination of data augmentation methods
Jun 12, 2020



Abstract:Purpose: This study aimed to develop and validate computer-aided diagnosis (CXDx) system for classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray (CXR) images. Materials and Methods: From two public datasets, 1248 CXR images were obtained, which included 215, 533, and 500 CXR images of COVID-19 pneumonia patients, non-COVID-19 pneumonia patients, and the healthy samples. The proposed CADx system utilized VGG16 as a pre-trained model and combination of conventional method and mixup as data augmentation methods. Other types of pre-trained models were compared with the VGG16-based model. Single type or no data augmentation methods were also evaluated. Splitting of training/validation/test sets was used when building and evaluating the CADx system. Three-category accuracy was evaluated for test set with 125 CXR images. Results: The three-category accuracy of the CAD system was 83.6% between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy. Sensitivity for COVID-19 pneumonia was more than 90%. The combination of conventional method and mixup was more useful than single type or no data augmentation method. Conclusion: This study was able to create an accurate CADx system for the 3-category classification. Source code of our CADx system is available as open source for COVID-19 research.
Automatic detection of acute ischemic stroke using non-contrast computed tomography and two-stage deep learning model
Apr 09, 2020



Abstract:Background and Purpose: We aimed to develop and evaluate an automatic acute ischemic stroke-related (AIS) detection system involving a two-stage deep learning model. Methods: We included 238 cases from two different institutions. AIS-related findings were annotated on each of the 238 sets of head CT images by referring to head magnetic resonance imaging (MRI) images in which an MRI examination was performed within 24 h following the CT scan. These 238 annotated cases were divided into a training set including 189 cases and test set including 49 cases. Subsequently, a two-stage deep learning detection model was constructed from the training set using the You Only Look Once v3 model and Visual Geometry Group 16 classification model. Then, the two-stage model performed the AIS detection process in the test set. To assess the detection model's results, a board-certified radiologist also evaluated the test set head CT images with and without the aid of the detection model. The sensitivity of AIS detection and number of false positives were calculated for the evaluation of the test set detection results. The sensitivity of the radiologist with and without the software detection results was compared using the McNemar test. A p-value of less than 0.05 was considered statistically significant. Results: For the two-stage model and radiologist without and with the use of the software results, the sensitivity was 37.3%, 33.3%, and 41.3%, respectively, and the number of false positives per one case was 1.265, 0.327, and 0.388, respectively. On using the two-stage detection model's results, the board-certified radiologist's detection sensitivity significantly improved (p-value = 0.0313). Conclusions: Our detection system involving the two-stage deep learning model significantly improved the radiologist's sensitivity in AIS detection.
Lung segmentation on chest x-ray images in patients with severe abnormal findings using deep learning
Aug 21, 2019



Abstract:Rationale and objectives: Several studies have evaluated the usefulness of deep learning for lung segmentation using chest x-ray (CXR) images with small- or medium-sized abnormal findings. Here, we built a database including both CXR images with severe abnormalities and experts' lung segmentation results, and aimed to evaluate our network's efficacy in lung segmentation from these images. Materials and Methods: For lung segmentation, CXR images from the Japanese Society of Radiological Technology (JSRT, N = 247) and Montgomery databases (N = 138), were included, and 65 additional images depicting severe abnormalities from a public database were evaluated and annotated by a radiologist, thereby adding lung segmentation results to these images. Baseline U-net was used to segment the lungs in images from the three databases. Subsequently, the U-net network architecture was automatically optimized for lung segmentation from CXR images using Bayesian optimization. Dice similarity coefficient (DSC) was calculated to confirm segmentation. Results: Our results demonstrated that using baseline U-net yielded poorer lung segmentation results in our database than those in the JSRT and Montgomery databases, implying that robust segmentation of lungs may be difficult because of severe abnormalities. The DSC values with baseline U-net for the JSRT, Montgomery and our databases were 0.979, 0.941, and 0.889, respectively, and with optimized U-net, 0.976, 0.973, and 0.932, respectively. Conclusion: For robust lung segmentation, the U-net architecture was optimized via Bayesian optimization, and our results demonstrate that the optimized U-net was more robust than baseline U-net in lung segmentation from CXR images with large-sized abnormalities.
Computer-aided diagnosis of lung nodule using gradient tree boosting and Bayesian optimization
Aug 28, 2017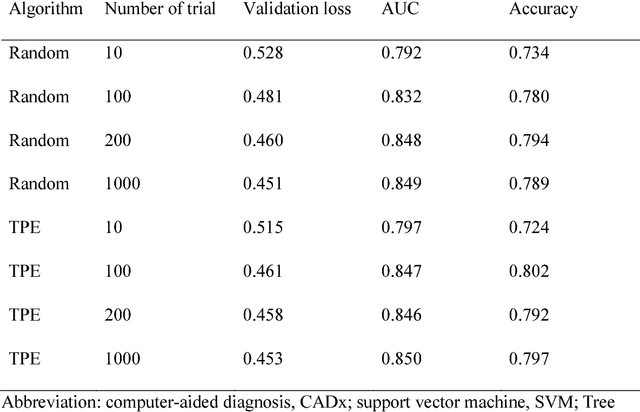
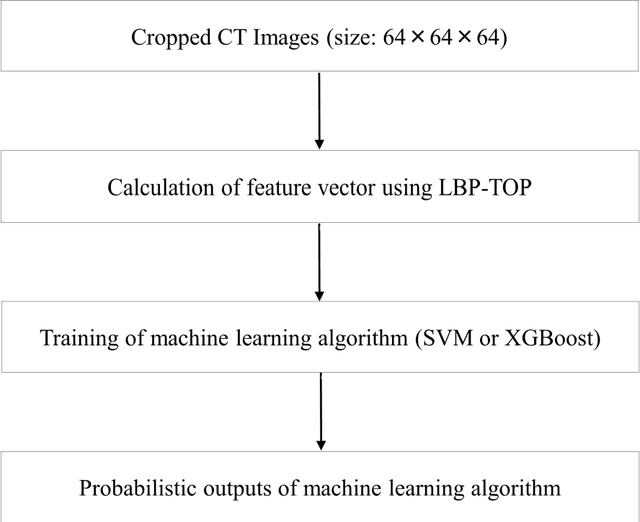
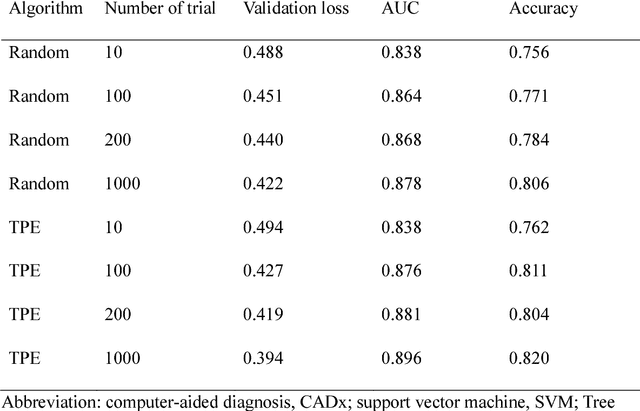
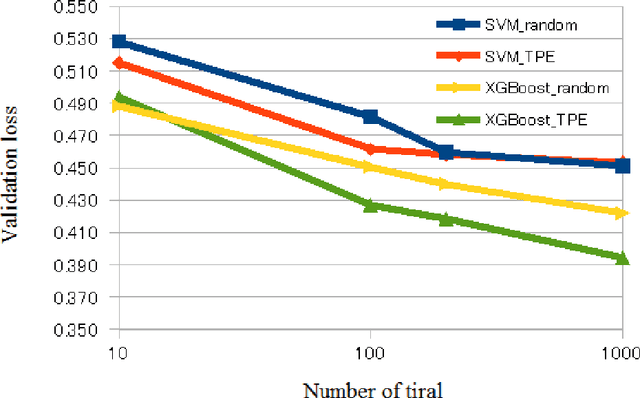
Abstract:We aimed to evaluate computer-aided diagnosis (CADx) system for lung nodule classification focusing on (i) usefulness of gradient tree boosting (XGBoost) and (ii) effectiveness of parameter optimization using Bayesian optimization (Tree Parzen Estimator, TPE) and random search. 99 lung nodules (62 lung cancers and 37 benign lung nodules) were included from public databases of CT images. A variant of local binary pattern was used for calculating feature vectors. Support vector machine (SVM) or XGBoost was trained using the feature vectors and their labels. TPE or random search was used for parameter optimization of SVM and XGBoost. Leave-one-out cross-validation was used for optimizing and evaluating the performance of our CADx system. Performance was evaluated using area under the curve (AUC) of receiver operating characteristic analysis. AUC was calculated 10 times, and its average was obtained. The best averaged AUC of SVM and XGBoost were 0.850 and 0.896, respectively; both were obtained using TPE. XGBoost was generally superior to SVM. Optimal parameters for achieving high AUC were obtained with fewer numbers of trials when using TPE, compared with random search. In conclusion, XGBoost was better than SVM for classifying lung nodules. TPE was more efficient than random search for parameter optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge