Ryosuke Kojima
Graduate School of Medicine, Kyoto University
Supervised Graph Contrastive Learning for Gene Regulatory Network
May 23, 2025Abstract:Graph representation learning is effective for obtaining a meaningful latent space utilizing the structure of graph data and is widely applied, including biological networks. In particular, Graph Contrastive Learning (GCL) has emerged as a powerful self-supervised method that relies on applying perturbations to graphs for data augmentation. However, when applying existing GCL methods to biological networks such as Gene Regulatory Networks (GRNs), they overlooked meaningful biologically relevant perturbations, e.g., gene knockdowns. In this study, we introduce SupGCL (Supervised Graph Contrastive Learning), a novel GCL method for GRNs that directly incorporates biological perturbations derived from gene knockdown experiments as the supervision. SupGCL mathematically extends existing GCL methods that utilize non-biological perturbations to probabilistic models that introduce actual biological gene perturbation utilizing gene knockdown data. Using the GRN representation obtained by our proposed method, our aim is to improve the performance of biological downstream tasks such as patient hazard prediction and disease subtype classification (graph-level task), and gene function classification (node-level task). We applied SupGCL on real GRN datasets derived from patients with multiple types of cancer, and in all experiments SupGCL achieves better performance than state-of-the-art baselines.
Learning Deep Dissipative Dynamics
Aug 21, 2024Abstract:This study challenges strictly guaranteeing ``dissipativity'' of a dynamical system represented by neural networks learned from given time-series data. Dissipativity is a crucial indicator for dynamical systems that generalizes stability and input-output stability, known to be valid across various systems including robotics, biological systems, and molecular dynamics. By analytically proving the general solution to the nonlinear Kalman-Yakubovich-Popov (KYP) lemma, which is the necessary and sufficient condition for dissipativity, we propose a differentiable projection that transforms any dynamics represented by neural networks into dissipative ones and a learning method for the transformed dynamics. Utilizing the generality of dissipativity, our method strictly guarantee stability, input-output stability, and energy conservation of trained dynamical systems. Finally, we demonstrate the robustness of our method against out-of-domain input through applications to robotic arms and fluid dynamics. Code here https://github.com/kojima-r/DeepDissipativeModel
Out-of-distribution Reject Option Method for Dataset Shift Problem in Early Disease Onset Prediction
May 30, 2024Abstract:Machine learning is increasingly used to predict lifestyle-related disease onset using health and medical data. However, the prediction effectiveness is hindered by dataset shift, which involves discrepancies in data distribution between the training and testing datasets, misclassifying out-of-distribution (OOD) data. To diminish dataset shift effects, this paper proposes the out-of-distribution reject option for prediction (ODROP), which integrates OOD detection models to preclude OOD data from the prediction phase. We investigated the efficacy of five OOD detection methods (variational autoencoder, neural network ensemble std, neural network ensemble epistemic, neural network energy, and neural network gaussian mixture based energy measurement) across two datasets, the Hirosaki and Wakayama health checkup data, in the context of three disease onset prediction tasks: diabetes, dyslipidemia, and hypertension. To evaluate the ODROP method, we trained disease onset prediction models and OOD detection models on Hirosaki data and used AUROC-rejection curve plots from Wakayama data. The variational autoencoder method showed superior stability and magnitude of improvement in Area Under the Receiver Operating Curve (AUROC) in five cases: AUROC in the Wakayama data was improved from 0.80 to 0.90 at a 31.1% rejection rate for diabetes onset and from 0.70 to 0.76 at a 34% rejection rate for dyslipidemia. We categorized dataset shifts into two types using SHAP clustering - those that considerably affect predictions and those that do not. We expect that this classification will help standardize measuring instruments. This study is the first to apply OOD detection to actual health and medical data, demonstrating its potential to substantially improve the accuracy and reliability of disease prediction models amidst dataset shift.
ReactionT5: a large-scale pre-trained model towards application of limited reaction data
Nov 12, 2023



Abstract:Transformer-based deep neural networks have revolutionized the field of molecular-related prediction tasks by treating molecules as symbolic sequences. These models have been successfully applied in various organic chemical applications by pretraining them with extensive compound libraries and subsequently fine-tuning them with smaller in-house datasets for specific tasks. However, many conventional methods primarily focus on single molecules, with limited exploration of pretraining for reactions involving multiple molecules. In this paper, we propose ReactionT5, a novel model that leverages pretraining on the Open Reaction Database (ORD), a publicly available large-scale resource. We further fine-tune this model for yield prediction and product prediction tasks, demonstrating its impressive performance even with limited fine-tuning data compared to traditional models. The pre-trained ReactionT5 model is publicly accessible on the Hugging Face platform.
A New Deep State-Space Analysis Framework for Patient Latent State Estimation and Classification from EHR Time Series Data
Jul 21, 2023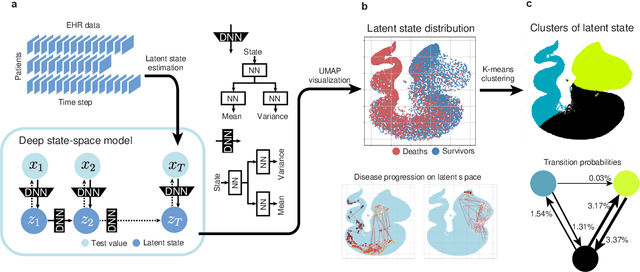
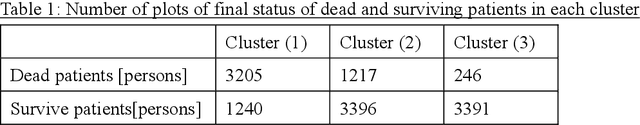
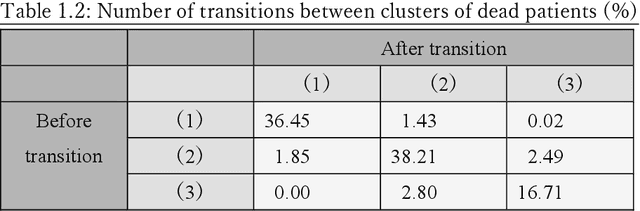
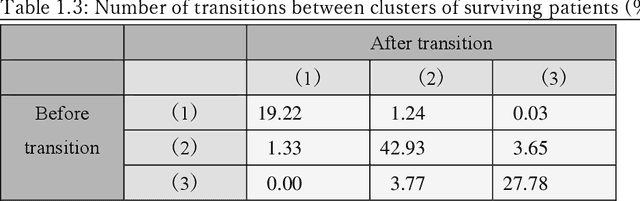
Abstract:Many diseases, including cancer and chronic conditions, require extended treatment periods and long-term strategies. Machine learning and AI research focusing on electronic health records (EHRs) have emerged to address this need. Effective treatment strategies involve more than capturing sequential changes in patient test values. It requires an explainable and clinically interpretable model by capturing the patient's internal state over time. In this study, we propose the "deep state-space analysis framework," using time-series unsupervised learning of EHRs with a deep state-space model. This framework enables learning, visualizing, and clustering of temporal changes in patient latent states related to disease progression. We evaluated our framework using time-series laboratory data from 12,695 cancer patients. By estimating latent states, we successfully discover latent states related to prognosis. By visualization and cluster analysis, the temporal transition of patient status and test items during state transitions characteristic of each anticancer drug were identified. Our framework surpasses existing methods in capturing interpretable latent space. It can be expected to enhance our comprehension of disease progression from EHRs, aiding treatment adjustments and prognostic determinations.
An end-to-end framework for gene expression classification by integrating a background knowledge graph: application to cancer prognosis prediction
Jun 29, 2023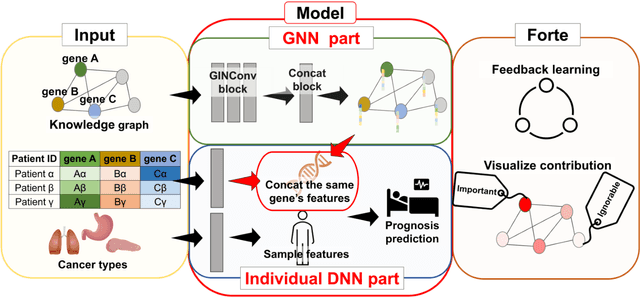
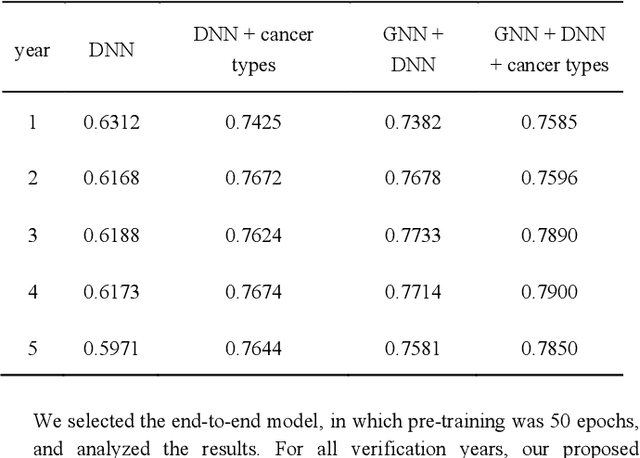
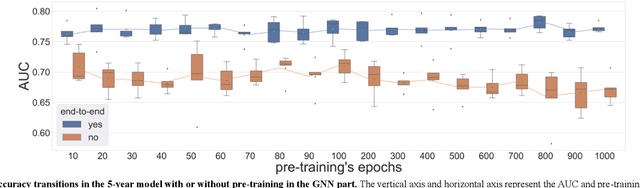
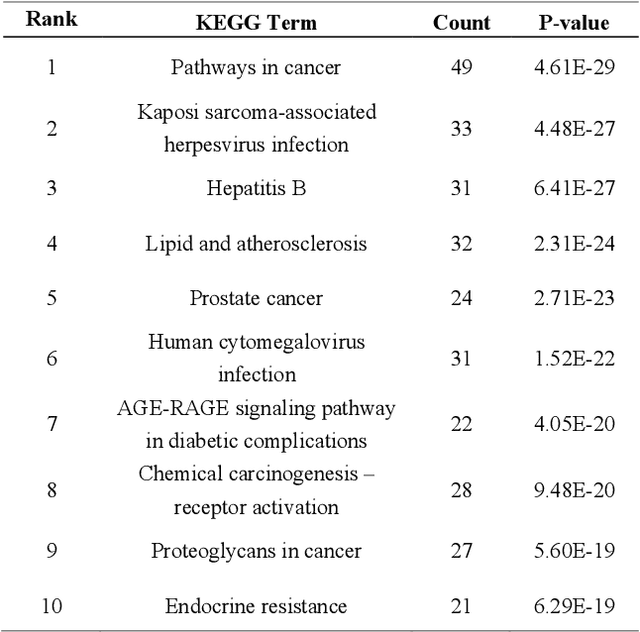
Abstract:Biological data may be separated into primary data, such as gene expression, and secondary data, such as pathways and protein-protein interactions. Methods using secondary data to enhance the analysis of primary data are promising, because secondary data have background information that is not included in primary data. In this study, we proposed an end-to-end framework to integrally handle secondary data to construct a classification model for primary data. We applied this framework to cancer prognosis prediction using gene expression data and a biological network. Cross-validation results indicated that our model achieved higher accuracy compared with a deep neural network model without background biological network information. Experiments conducted in patient groups by cancer type showed improvement in ROC-area under the curve for many groups. Visualizations of high accuracy cancer types identified contributing genes and pathways by enrichment analysis. Known biomarkers and novel biomarker candidates were identified through these experiments.
GraphIX: Graph-based In silico XAI for drug repositioning from biopharmaceutical network
Dec 27, 2022Abstract:Drug repositioning holds great promise because it can reduce the time and cost of new drug development. While drug repositioning can omit various R&D processes, confirming pharmacological effects on biomolecules is essential for application to new diseases. Biomedical explainability in a drug repositioning model can support appropriate insights in subsequent in-depth studies. However, the validity of the XAI methodology is still under debate, and the effectiveness of XAI in drug repositioning prediction applications remains unclear. In this study, we propose GraphIX, an explainable drug repositioning framework using biological networks, and quantitatively evaluate its explainability. GraphIX first learns the network weights and node features using a graph neural network from known drug indication and knowledge graph that consists of three types of nodes (but not given node type information): disease, drug, and protein. Analysis of the post-learning features showed that node types that were not known to the model beforehand are distinguished through the learning process based on the graph structure. From the learned weights and features, GraphIX then predicts the disease-drug association and calculates the contribution values of the nodes located in the neighborhood of the predicted disease and drug. We hypothesized that the neighboring protein node to which the model gave a high contribution is important in understanding the actual pharmacological effects. Quantitative evaluation of the validity of protein nodes' contribution using a real-world database showed that the high contribution proteins shown by GraphIX are reasonable as a mechanism of drug action. GraphIX is a framework for evidence-based drug discovery that can present to users new disease-drug associations and identify the protein important for understanding its pharmacological effects from a large and complex knowledge base.
Learning Deep Input-Output Stable Dynamics
Jun 27, 2022



Abstract:Learning stable dynamics from observed time-series data is an essential problem in robotics, physical modeling, and systems biology. Many of these dynamics are represented as an inputs-output system to communicate with the external environment. In this study, we focus on input-output stable systems, exhibiting robustness against unexpected stimuli and noise. We propose a method to learn nonlinear systems guaranteeing the input-output stability. Our proposed method utilizes the differentiable projection onto the space satisfying the Hamilton-Jacobi inequality to realize the input-output stability. The problem of finding this projection can be formulated as a quadratic constraint quadratic programming problem, and we derive the particular solution analytically. Also, we apply our method to a toy bistable model and the task of training a benchmark generated from a glucose-insulin simulator. The results show that the nonlinear system with neural networks by our method achieves the input-output stability, unlike naive neural networks. Our code is available at https://github.com/clinfo/DeepIOStability.
Individual health-disease phase diagrams for disease prevention based on machine learning
May 31, 2022



Abstract:Early disease detection and prevention methods based on effective interventions are gaining attention. Machine learning technology has enabled precise disease prediction by capturing individual differences in multivariate data. Progress in precision medicine has revealed that substantial heterogeneity exists in health data at the individual level and that complex health factors are involved in the development of chronic diseases. However, it remains a challenge to identify individual physiological state changes in cross-disease onset processes because of the complex relationships among multiple biomarkers. Here, we present the health-disease phase diagram (HDPD), which represents a personal health state by visualizing the boundary values of multiple biomarkers that fluctuate early in the disease progression process. In HDPDs, future onset predictions are represented by perturbing multiple biomarker values while accounting for dependencies among variables. We constructed HDPDs for 11 non-communicable diseases (NCDs) from a longitudinal health checkup cohort of 3,238 individuals, comprising 3,215 measurement items and genetic data. Improvement of biomarker values to the non-onset region in HDPD significantly prevented future disease onset in 7 out of 11 NCDs. Our results demonstrate that HDPDs can represent individual physiological states in the onset process and be used as intervention goals for disease prevention.
MatSat: a matrix-based differentiable SAT solver
Aug 14, 2021

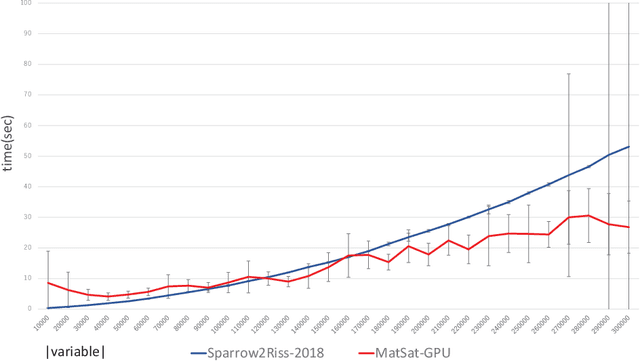
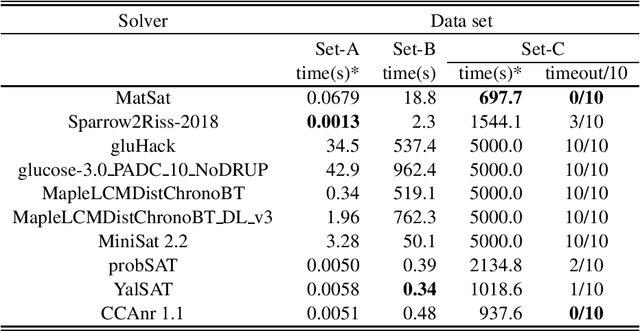
Abstract:We propose a new approach to SAT solving which solves SAT problems in vector spaces as a cost minimization problem of a non-negative differentiable cost function J^sat. In our approach, a solution, i.e., satisfying assignment, for a SAT problem in n variables is represented by a binary vector u in {0,1}^n that makes J^sat(u) zero. We search for such u in a vector space R^n by cost minimization, i.e., starting from an initial u_0 and minimizing J to zero while iteratively updating u by Newton's method. We implemented our approach as a matrix-based differential SAT solver MatSat. Although existing main-stream SAT solvers decide each bit of a solution assignment one by one, be they of conflict driven clause learning (CDCL) type or of stochastic local search (SLS) type, MatSat fundamentally differs from them in that it continuously approach a solution in a vector space. We conducted an experiment to measure the scalability of MatSat with random 3-SAT problems in which MatSat could find a solution up to n=10^5 variables. We also compared MatSat with four state-of-the-art SAT solvers including winners of SAT competition 2018 and SAT Race 2019 in terms of time for finding a solution, using a random benchmark set from SAT 2018 competition and an artificial random 3-SAT instance set. The result shows that MatSat comes in second in both test sets and outperforms all the CDCL type solvers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge