Yohei Mineharu
Out-of-distribution Reject Option Method for Dataset Shift Problem in Early Disease Onset Prediction
May 30, 2024Abstract:Machine learning is increasingly used to predict lifestyle-related disease onset using health and medical data. However, the prediction effectiveness is hindered by dataset shift, which involves discrepancies in data distribution between the training and testing datasets, misclassifying out-of-distribution (OOD) data. To diminish dataset shift effects, this paper proposes the out-of-distribution reject option for prediction (ODROP), which integrates OOD detection models to preclude OOD data from the prediction phase. We investigated the efficacy of five OOD detection methods (variational autoencoder, neural network ensemble std, neural network ensemble epistemic, neural network energy, and neural network gaussian mixture based energy measurement) across two datasets, the Hirosaki and Wakayama health checkup data, in the context of three disease onset prediction tasks: diabetes, dyslipidemia, and hypertension. To evaluate the ODROP method, we trained disease onset prediction models and OOD detection models on Hirosaki data and used AUROC-rejection curve plots from Wakayama data. The variational autoencoder method showed superior stability and magnitude of improvement in Area Under the Receiver Operating Curve (AUROC) in five cases: AUROC in the Wakayama data was improved from 0.80 to 0.90 at a 31.1% rejection rate for diabetes onset and from 0.70 to 0.76 at a 34% rejection rate for dyslipidemia. We categorized dataset shifts into two types using SHAP clustering - those that considerably affect predictions and those that do not. We expect that this classification will help standardize measuring instruments. This study is the first to apply OOD detection to actual health and medical data, demonstrating its potential to substantially improve the accuracy and reliability of disease prediction models amidst dataset shift.
A New Deep State-Space Analysis Framework for Patient Latent State Estimation and Classification from EHR Time Series Data
Jul 21, 2023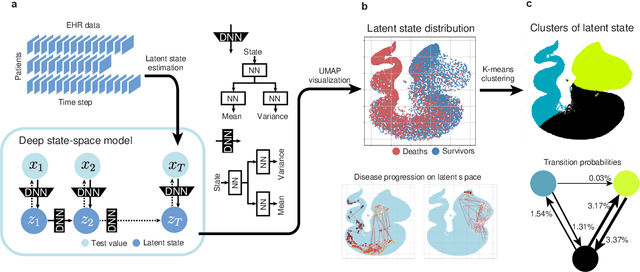
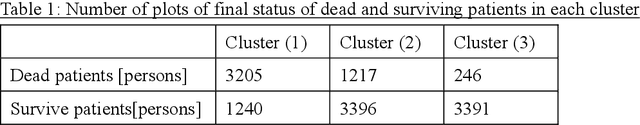
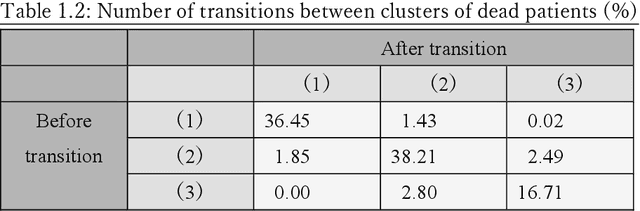
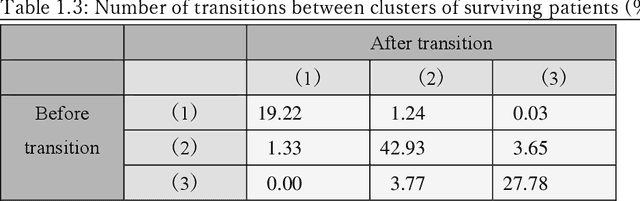
Abstract:Many diseases, including cancer and chronic conditions, require extended treatment periods and long-term strategies. Machine learning and AI research focusing on electronic health records (EHRs) have emerged to address this need. Effective treatment strategies involve more than capturing sequential changes in patient test values. It requires an explainable and clinically interpretable model by capturing the patient's internal state over time. In this study, we propose the "deep state-space analysis framework," using time-series unsupervised learning of EHRs with a deep state-space model. This framework enables learning, visualizing, and clustering of temporal changes in patient latent states related to disease progression. We evaluated our framework using time-series laboratory data from 12,695 cancer patients. By estimating latent states, we successfully discover latent states related to prognosis. By visualization and cluster analysis, the temporal transition of patient status and test items during state transitions characteristic of each anticancer drug were identified. Our framework surpasses existing methods in capturing interpretable latent space. It can be expected to enhance our comprehension of disease progression from EHRs, aiding treatment adjustments and prognostic determinations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge