Kaori Togashi
Lung segmentation on chest x-ray images in patients with severe abnormal findings using deep learning
Aug 21, 2019



Abstract:Rationale and objectives: Several studies have evaluated the usefulness of deep learning for lung segmentation using chest x-ray (CXR) images with small- or medium-sized abnormal findings. Here, we built a database including both CXR images with severe abnormalities and experts' lung segmentation results, and aimed to evaluate our network's efficacy in lung segmentation from these images. Materials and Methods: For lung segmentation, CXR images from the Japanese Society of Radiological Technology (JSRT, N = 247) and Montgomery databases (N = 138), were included, and 65 additional images depicting severe abnormalities from a public database were evaluated and annotated by a radiologist, thereby adding lung segmentation results to these images. Baseline U-net was used to segment the lungs in images from the three databases. Subsequently, the U-net network architecture was automatically optimized for lung segmentation from CXR images using Bayesian optimization. Dice similarity coefficient (DSC) was calculated to confirm segmentation. Results: Our results demonstrated that using baseline U-net yielded poorer lung segmentation results in our database than those in the JSRT and Montgomery databases, implying that robust segmentation of lungs may be difficult because of severe abnormalities. The DSC values with baseline U-net for the JSRT, Montgomery and our databases were 0.979, 0.941, and 0.889, respectively, and with optimized U-net, 0.976, 0.973, and 0.932, respectively. Conclusion: For robust lung segmentation, the U-net architecture was optimized via Bayesian optimization, and our results demonstrate that the optimized U-net was more robust than baseline U-net in lung segmentation from CXR images with large-sized abnormalities.
Computer-aided diagnosis of lung nodule using gradient tree boosting and Bayesian optimization
Aug 28, 2017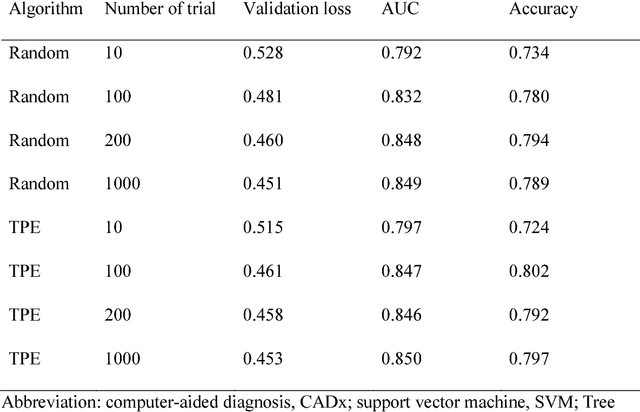
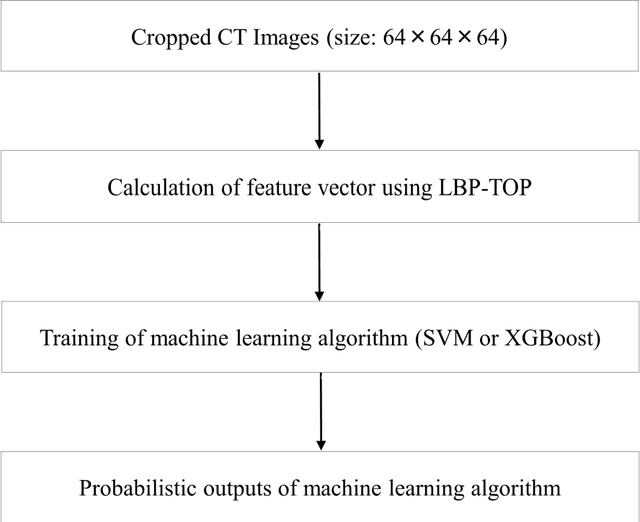
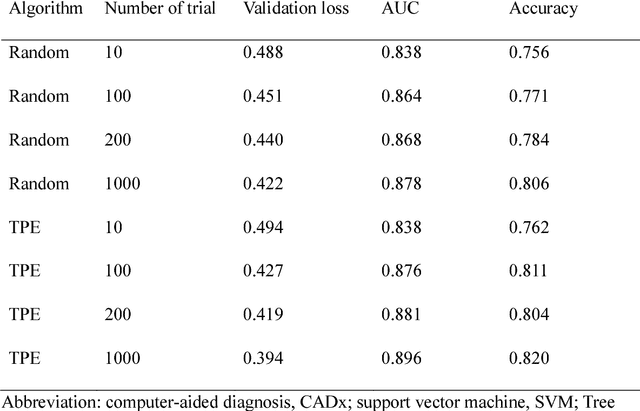
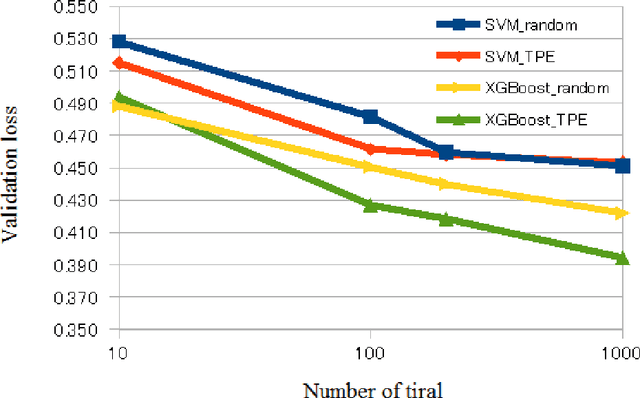
Abstract:We aimed to evaluate computer-aided diagnosis (CADx) system for lung nodule classification focusing on (i) usefulness of gradient tree boosting (XGBoost) and (ii) effectiveness of parameter optimization using Bayesian optimization (Tree Parzen Estimator, TPE) and random search. 99 lung nodules (62 lung cancers and 37 benign lung nodules) were included from public databases of CT images. A variant of local binary pattern was used for calculating feature vectors. Support vector machine (SVM) or XGBoost was trained using the feature vectors and their labels. TPE or random search was used for parameter optimization of SVM and XGBoost. Leave-one-out cross-validation was used for optimizing and evaluating the performance of our CADx system. Performance was evaluated using area under the curve (AUC) of receiver operating characteristic analysis. AUC was calculated 10 times, and its average was obtained. The best averaged AUC of SVM and XGBoost were 0.850 and 0.896, respectively; both were obtained using TPE. XGBoost was generally superior to SVM. Optimal parameters for achieving high AUC were obtained with fewer numbers of trials when using TPE, compared with random search. In conclusion, XGBoost was better than SVM for classifying lung nodules. TPE was more efficient than random search for parameter optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge