Hoda Memarzadeh
Applying unsupervised keyphrase methods on concepts extracted from discharge sheets
Mar 15, 2023Abstract:Clinical notes containing valuable patient information are written by different health care providers with various scientific levels and writing styles. It might be helpful for clinicians and researchers to understand what information is essential when dealing with extensive electronic medical records. Entities recognizing and mapping them to standard terminologies is crucial in reducing ambiguity in processing clinical notes. Although named entity recognition and entity linking are critical steps in clinical natural language processing, they can also result in the production of repetitive and low-value concepts. In other hand, all parts of a clinical text do not share the same importance or content in predicting the patient's condition. As a result, it is necessary to identify the section in which each content is recorded and also to identify key concepts to extract meaning from clinical texts. In this study, these challenges have been addressed by using clinical natural language processing techniques. In addition, in order to identify key concepts, a set of popular unsupervised key phrase extraction methods has been verified and evaluated. Considering that most of the clinical concepts are in the form of multi-word expressions and their accurate identification requires the user to specify n-gram range, we have proposed a shortcut method to preserve the structure of the expression based on TF-IDF. In order to evaluate the pre-processing method and select the concepts, we have designed two types of downstream tasks (multiple and binary classification) using the capabilities of transformer-based models. The obtained results show the superiority of proposed method in combination with SciBERT model, also offer an insight into the efficacy of general extracting essential phrase methods for clinical notes.
Assessing mortality prediction through different representation models based on concepts extracted from clinical notes
Jul 22, 2022



Abstract:Recent years have seen particular interest in using electronic medical records (EMRs) for secondary purposes to enhance the quality and safety of healthcare delivery. EMRs tend to contain large amounts of valuable clinical notes. Learning of embedding is a method for converting notes into a format that makes them comparable. Transformer-based representation models have recently made a great leap forward. These models are pre-trained on large online datasets to understand natural language texts effectively. The quality of a learning embedding is influenced by how clinical notes are used as input to representation models. A clinical note has several sections with different levels of information value. It is also common for healthcare providers to use different expressions for the same concept. Existing methods use clinical notes directly or with an initial preprocessing as input to representation models. However, to learn a good embedding, we identified the most essential clinical notes section. We then mapped the extracted concepts from selected sections to the standard names in the Unified Medical Language System (UMLS). We used the standard phrases corresponding to the unique concepts as input for clinical models. We performed experiments to measure the usefulness of the learned embedding vectors in the task of hospital mortality prediction on a subset of the publicly available Medical Information Mart for Intensive Care (MIMIC-III) dataset. According to the experiments, clinical transformer-based representation models produced better results with getting input generated by standard names of extracted unique concepts compared to other input formats. The best-performing models were BioBERT, PubMedBERT, and UmlsBERT, respectively.
Heterogeneous electronic medical record representation for similarity computing
Apr 29, 2021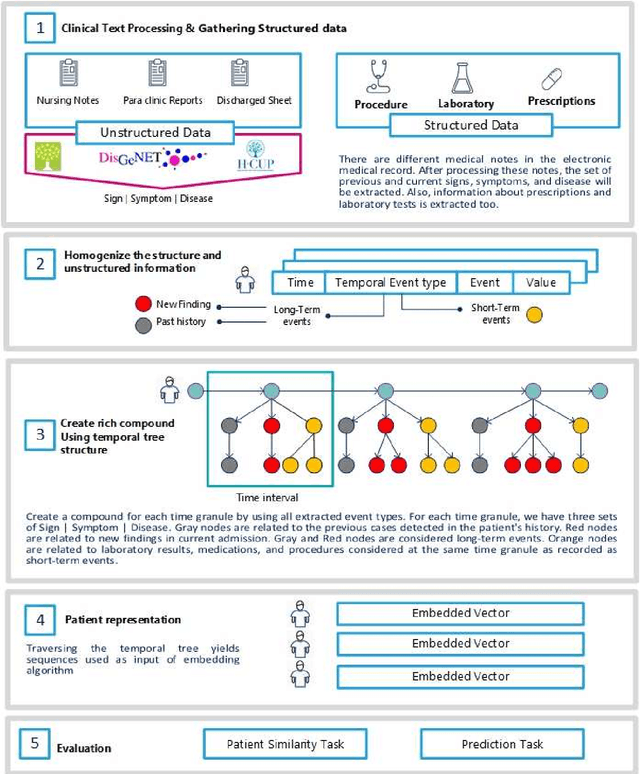
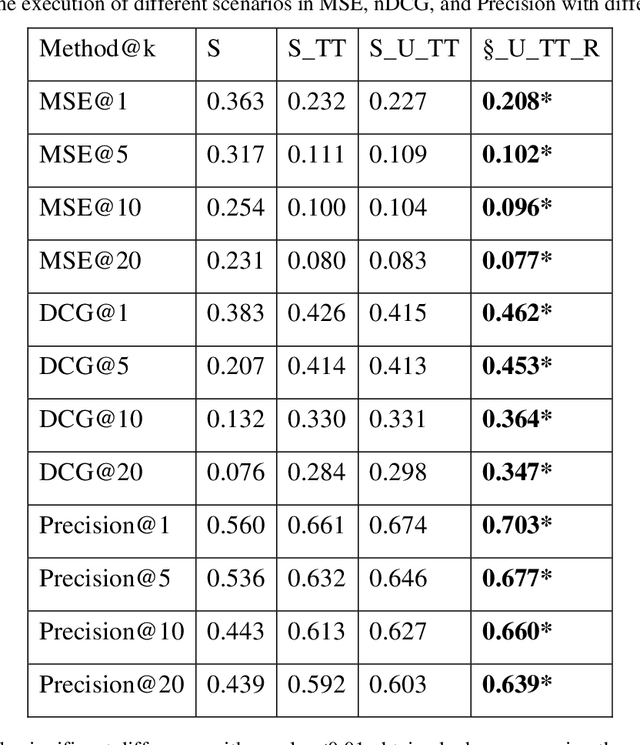


Abstract:Due to the widespread use of tools and the development of text processing techniques, the size and range of clinical data are not limited to structured data. The rapid growth of recorded information has led to big data platforms in healthcare that could be used to improve patients' primary care and serve various secondary purposes. Patient similarity assessment is one of the secondary tasks in identifying patients who are similar to a given patient, and it helps derive insights from similar patients' records to provide better treatment. This type of assessment is based on calculating the distance between patients. Since representing and calculating the similarity of patients plays an essential role in many secondary uses of electronic records, this article examines a new data representation method for Electronic Medical Records (EMRs) while taking into account the information in clinical narratives for similarity computing. Some previous works are based on structured data types, while other works only use unstructured data. However, a comprehensive representation of the information contained in the EMR requires the effective aggregation of both structured and unstructured data. To address the limitations of previous methods, we propose a method that captures the co-occurrence of different medical events, including signs, symptoms, and diseases extracted via unstructured data and structured data. It integrates data as discriminative features to construct a temporal tree, considering the difference between events that have short-term and long-term impacts. Our results show that considering signs, symptoms, and diseases in every time interval leads to less MSE and more precision compared to baseline representations that do not consider this information or consider them separately from structured data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge