Hendrik Burwinkel
U-PET: MRI-based Dementia Detection with Joint Generation of Synthetic FDG-PET Images
Jun 16, 2022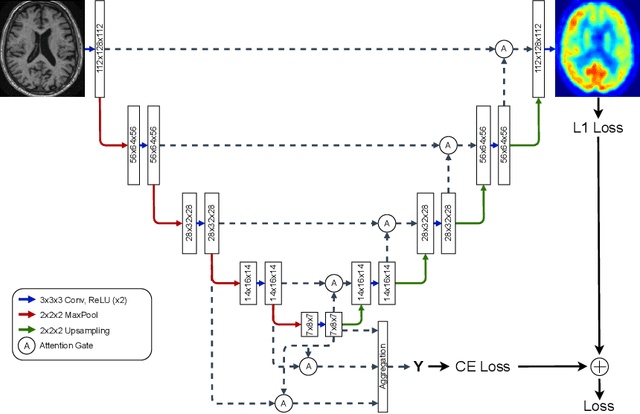
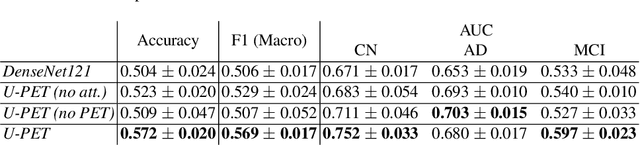
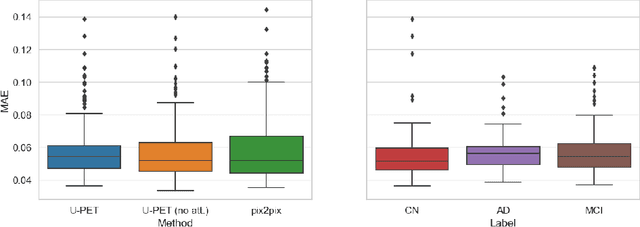
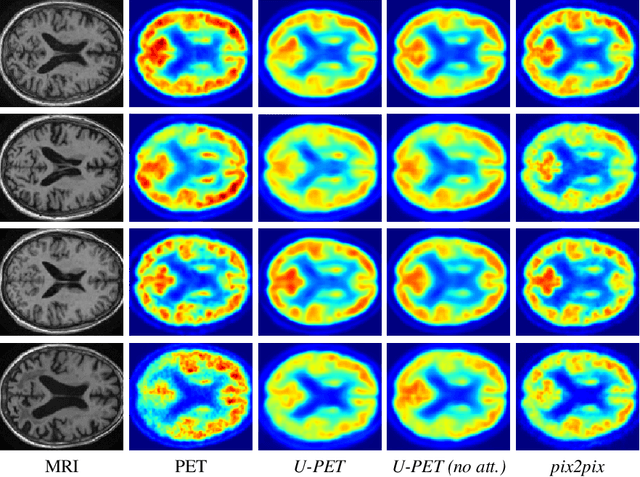
Abstract:Alzheimer's disease (AD) is the most common cause of dementia. An early detection is crucial for slowing down the disease and mitigating risks related to the progression. While the combination of MRI and FDG-PET is the best image-based tool for diagnosis, FDG-PET is not always available. The reliable detection of Alzheimer's disease with only MRI could be beneficial, especially in regions where FDG-PET might not be affordable for all patients. To this end, we propose a multi-task method based on U-Net that takes T1-weighted MR images as an input to generate synthetic FDG-PET images and classifies the dementia progression of the patient into cognitive normal (CN), cognitive impairment (MCI), and AD. The attention gates used in both task heads can visualize the most relevant parts of the brain, guiding the examiner and adding interpretability. Results show the successful generation of synthetic FDG-PET images and a performance increase in disease classification over the naive single-task baseline.
U-GAT: Multimodal Graph Attention Network for COVID-19 Outcome Prediction
Jul 29, 2021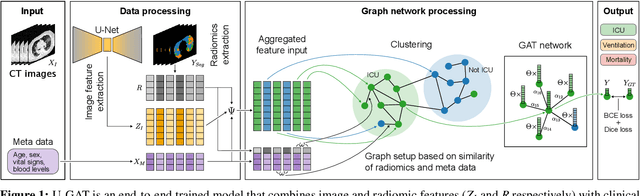
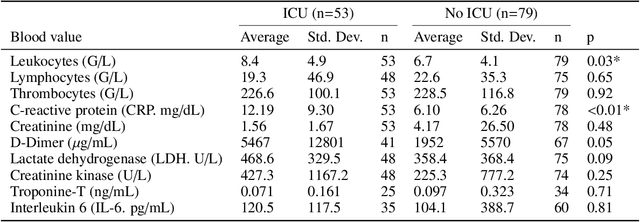
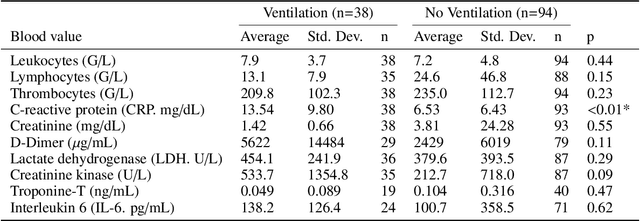
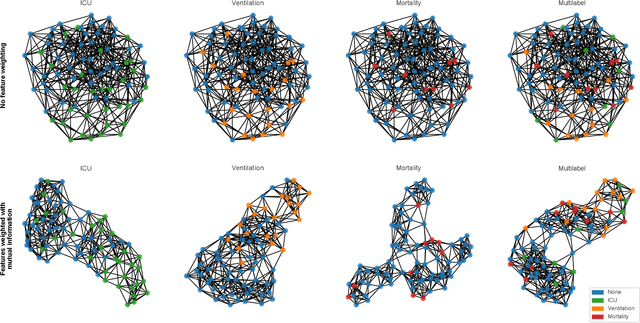
Abstract:During the first wave of COVID-19, hospitals were overwhelmed with the high number of admitted patients. An accurate prediction of the most likely individual disease progression can improve the planning of limited resources and finding the optimal treatment for patients. However, when dealing with a newly emerging disease such as COVID-19, the impact of patient- and disease-specific factors (e.g. body weight or known co-morbidities) on the immediate course of disease is by and large unknown. In the case of COVID-19, the need for intensive care unit (ICU) admission of pneumonia patients is often determined only by acute indicators such as vital signs (e.g. breathing rate, blood oxygen levels), whereas statistical analysis and decision support systems that integrate all of the available data could enable an earlier prognosis. To this end, we propose a holistic graph-based approach combining both imaging and non-imaging information. Specifically, we introduce a multimodal similarity metric to build a population graph for clustering patients and an image-based end-to-end Graph Attention Network to process this graph and predict the COVID-19 patient outcomes: admission to ICU, need for ventilation and mortality. Additionally, the network segments chest CT images as an auxiliary task and extracts image features and radiomics for feature fusion with the available metadata. Results on a dataset collected in Klinikum rechts der Isar in Munich, Germany show that our approach outperforms single modality and non-graph baselines. Moreover, our clustering and graph attention allow for increased understanding of the patient relationships within the population graph and provide insight into the network's decision-making process.
Simultaneous imputation and disease classification in incomplete medical datasets using Multigraph Geometric Matrix Completion (MGMC)
May 14, 2020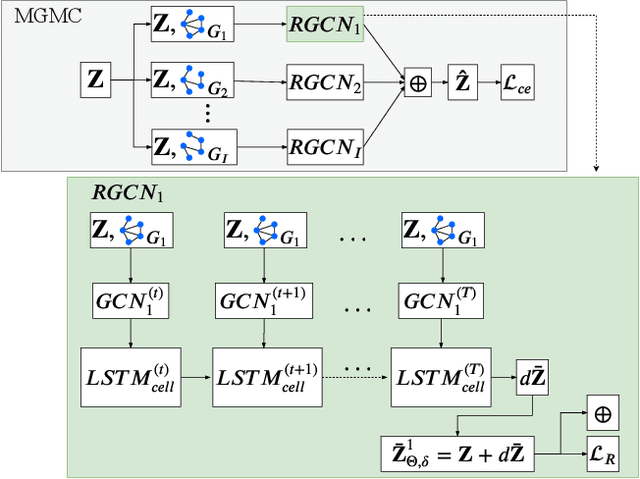
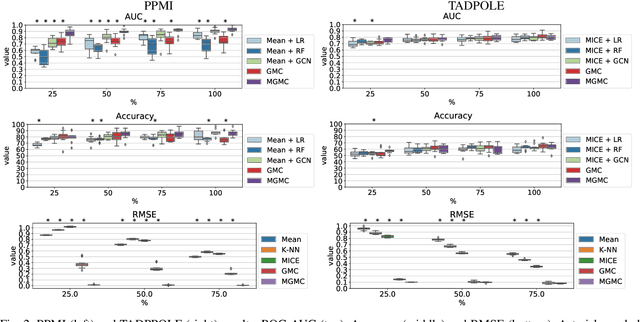
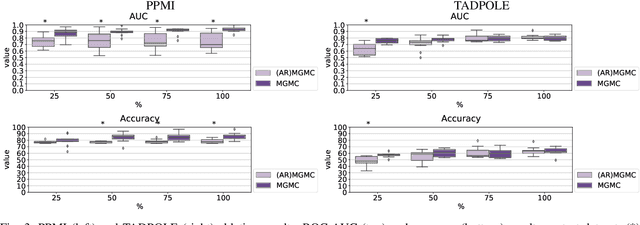
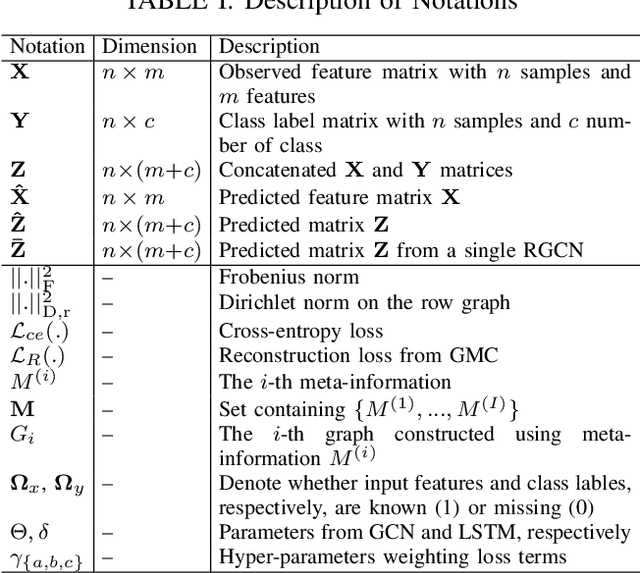
Abstract:Large-scale population-based studies in medicine are a key resource towards better diagnosis, monitoring, and treatment of diseases. They also serve as enablers of clinical decision support systems, in particular Computer Aided Diagnosis (CADx) using machine learning (ML). Numerous ML approaches for CADx have been proposed in literature. However, these approaches assume full data availability, which is not always feasible in clinical data. To account for missing data, incomplete data samples are either removed or imputed, which could lead to data bias and may negatively affect classification performance. As a solution, we propose an end-to-end learning of imputation and disease prediction of incomplete medical datasets via Multigraph Geometric Matrix Completion (MGMC). MGMC uses multiple recurrent graph convolutional networks, where each graph represents an independent population model based on a key clinical meta-feature like age, sex, or cognitive function. Graph signal aggregation from local patient neighborhoods, combined with multigraph signal fusion via self-attention, has a regularizing effect on both matrix reconstruction and classification performance. Our proposed approach is able to impute class relevant features as well as perform accurate classification on two publicly available medical datasets. We empirically show the superiority of our proposed approach in terms of classification and imputation performance when compared with state-of-the-art approaches. MGMC enables disease prediction in multimodal and incomplete medical datasets. These findings could serve as baseline for future CADx approaches which utilize incomplete datasets.
Domain-specific loss design for unsupervised physical training: A new approach to modeling medical ML solutions
May 09, 2020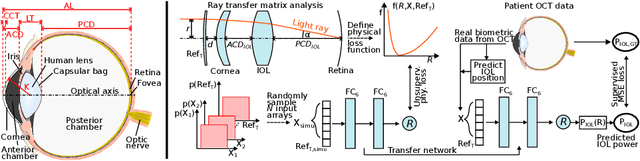
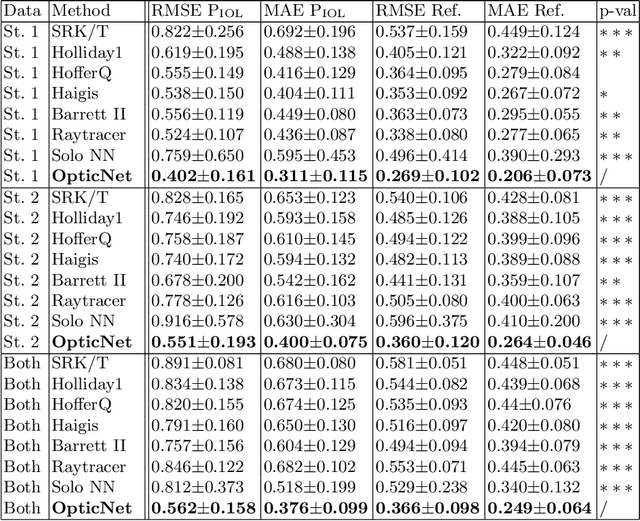
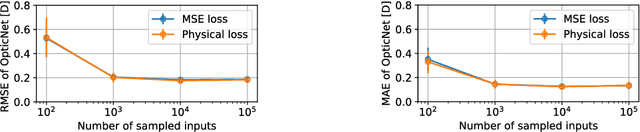
Abstract:Today, cataract surgery is the most frequently performed ophthalmic surgery in the world. The cataract, a developing opacity of the human eye lens, constitutes the world's most frequent cause for blindness. During surgery, the lens is removed and replaced by an artificial intraocular lens (IOL). To prevent patients from needing strong visual aids after surgery, a precise prediction of the optical properties of the inserted IOL is crucial. There has been lots of activity towards developing methods to predict these properties from biometric eye data obtained by OCT devices, recently also by employing machine learning. They consider either only biometric data or physical models, but rarely both, and often neglect the IOL geometry. In this work, we propose OpticNet, a novel optical refraction network, loss function, and training scheme which is unsupervised, domain-specific, and physically motivated. We derive a precise light propagation eye model using single-ray raytracing and formulate a differentiable loss function that back-propagates physical gradients into the network. Further, we propose a new transfer learning procedure, which allows unsupervised training on the physical model and fine-tuning of the network on a cohort of real IOL patient cases. We show that our network is not only superior to systems trained with standard procedures but also that our method outperforms the current state of the art in IOL calculation when compared on two biometric data sets.
Decision Support for Intoxication Prediction Using Graph Convolutional Networks
May 02, 2020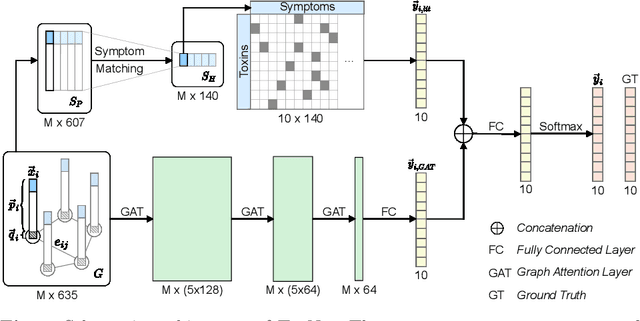
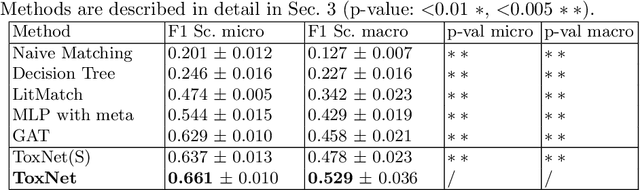
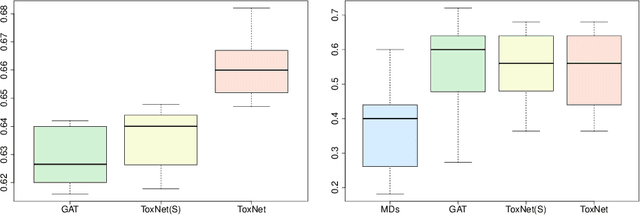
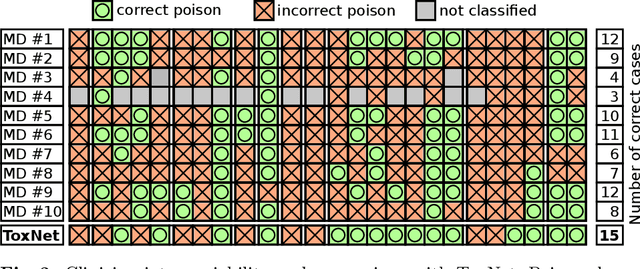
Abstract:Every day, poison control centers (PCC) are called for immediate classification and treatment recommendations if an acute intoxication is suspected. Due to the time-sensitive nature of these cases, doctors are required to propose a correct diagnosis and intervention within a minimal time frame. Usually the toxin is known and recommendations can be made accordingly. However, in challenging cases only symptoms are mentioned and doctors have to rely on their clinical experience. Medical experts and our analyses of a regional dataset of intoxication records provide evidence that this is challenging, since occurring symptoms may not always match the textbook description due to regional distinctions, inter-rater variance, and institutional workflow. Computer-aided diagnosis (CADx) can provide decision support, but approaches so far do not consider additional information of the reported cases like age or gender, despite their potential value towards a correct diagnosis. In this work, we propose a new machine learning based CADx method which fuses symptoms and meta information of the patients using graph convolutional networks. We further propose a novel symptom matching method that allows the effective incorporation of prior knowledge into the learning process and evidently stabilizes the poison prediction. We validate our method against 10 medical doctors with different experience diagnosing intoxication cases for 10 different toxins from the PCC in Munich and show our method's superiority in performance for poison prediction.
Multi-modal Graph Fusion for Inductive Disease Classification in Incomplete Datasets
May 08, 2019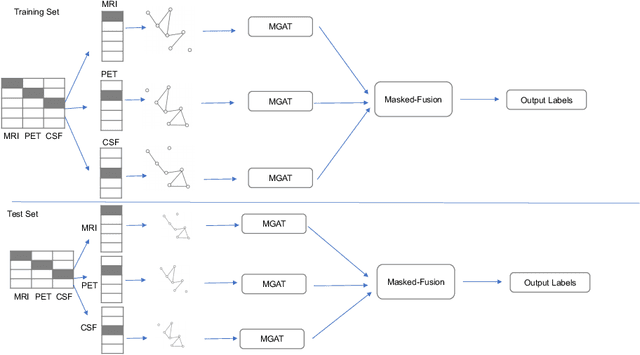
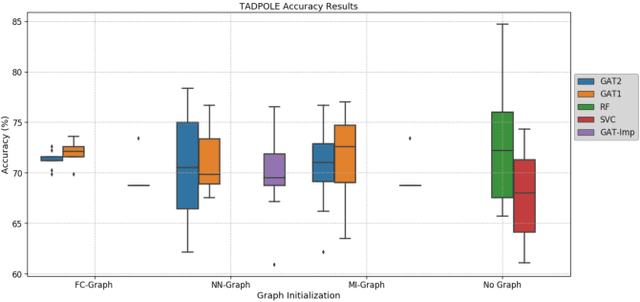
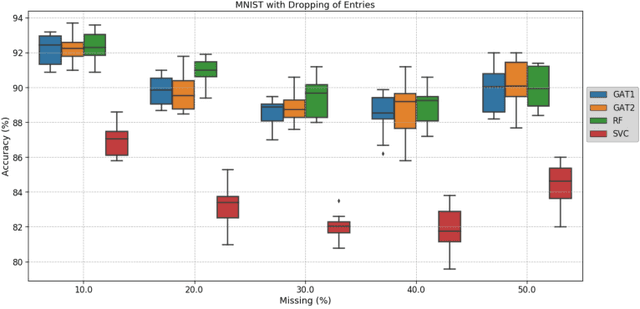
Abstract:Clinical diagnostic decision making and population-based studies often rely on multi-modal data which is noisy and incomplete. Recently, several works proposed geometric deep learning approaches to solve disease classification, by modeling patients as nodes in a graph, along with graph signal processing of multi-modal features. Many of these approaches are limited by assuming modality- and feature-completeness, and by transductive inference, which requires re-training of the entire model for each new test sample. In this work, we propose a novel inductive graph-based approach that can generalize to out-of-sample patients, despite missing features from entire modalities per patient. We propose multi-modal graph fusion which is trained end-to-end towards node-level classification. We demonstrate the fundamental working principle of this method on a simplified MNIST toy dataset. In experiments on medical data, our method outperforms single static graph approach in multi-modal disease classification.
Adaptive image-feature learning for disease classification using inductive graph networks
May 08, 2019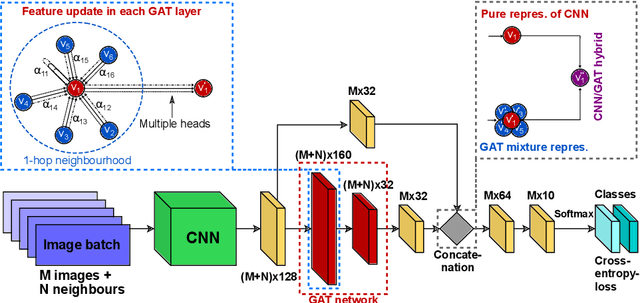
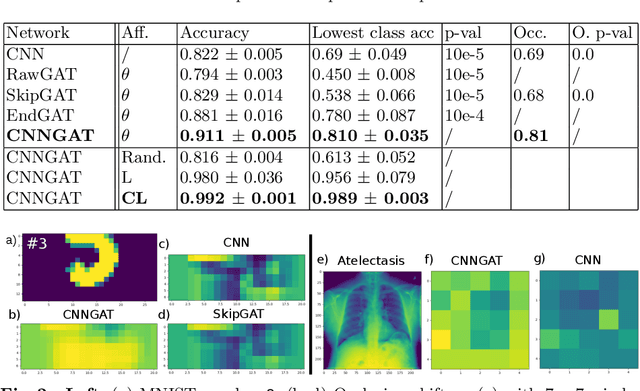
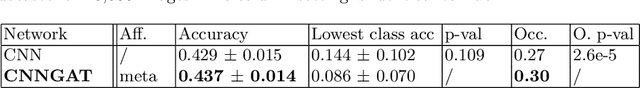
Abstract:Recently, Geometric Deep Learning (GDL) has been introduced as a novel and versatile framework for computer-aided disease classification. GDL uses patient meta-information such as age and gender to model patient cohort relations in a graph structure. Concepts from graph signal processing are leveraged to learn the optimal mapping of multi-modal features, e.g. from images to disease classes. Related studies so far have considered image features that are extracted in a pre-processing step. We hypothesize that such an approach prevents the network from optimizing feature representations towards achieving the best performance in the graph network. We propose a new network architecture that exploits an inductive end-to-end learning approach for disease classification, where filters from both the CNN and the graph are trained jointly. We validate this architecture against state-of-the-art inductive graph networks and demonstrate significantly improved classification scores on a modified MNIST toy dataset, as well as comparable classification results with higher stability on a chest X-ray image dataset. Additionally, we explain how the structural information of the graph affects both the image filters and the feature learning.
InceptionGCN: Receptive Field Aware Graph Convolutional Network for Disease Prediction
Mar 11, 2019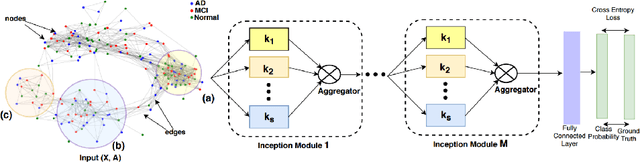
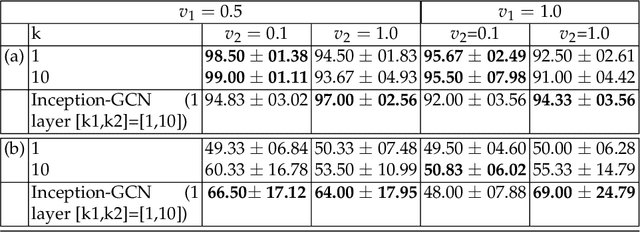
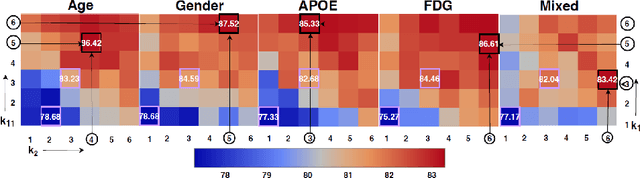
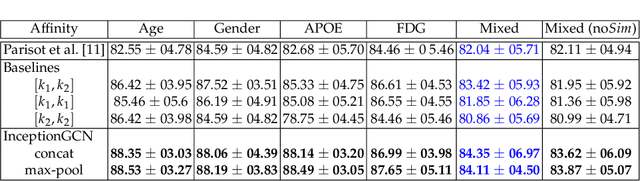
Abstract:Geometric deep learning provides a principled and versatile manner for the integration of imaging and non-imaging modalities in the medical domain. Graph Convolutional Networks (GCNs) in particular have been explored on a wide variety of problems such as disease prediction, segmentation, and matrix completion by leveraging large, multimodal datasets. In this paper, we introduce a new spectral domain architecture for deep learning on graphs for disease prediction. The novelty lies in defining geometric 'inception modules' which are capable of capturing intra- and inter-graph structural heterogeneity during convolutions. We design filters with different kernel sizes to build our architecture. We show our disease prediction results on two publicly available datasets. Further, we provide insights on the behaviour of regular GCNs and our proposed model under varying input scenarios on simulated data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge