Marcus R. Makowski
Biomedical Large Languages Models Seem not to be Superior to Generalist Models on Unseen Medical Data
Aug 25, 2024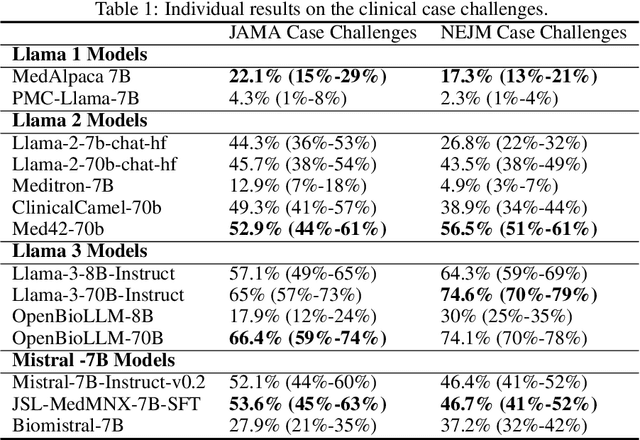
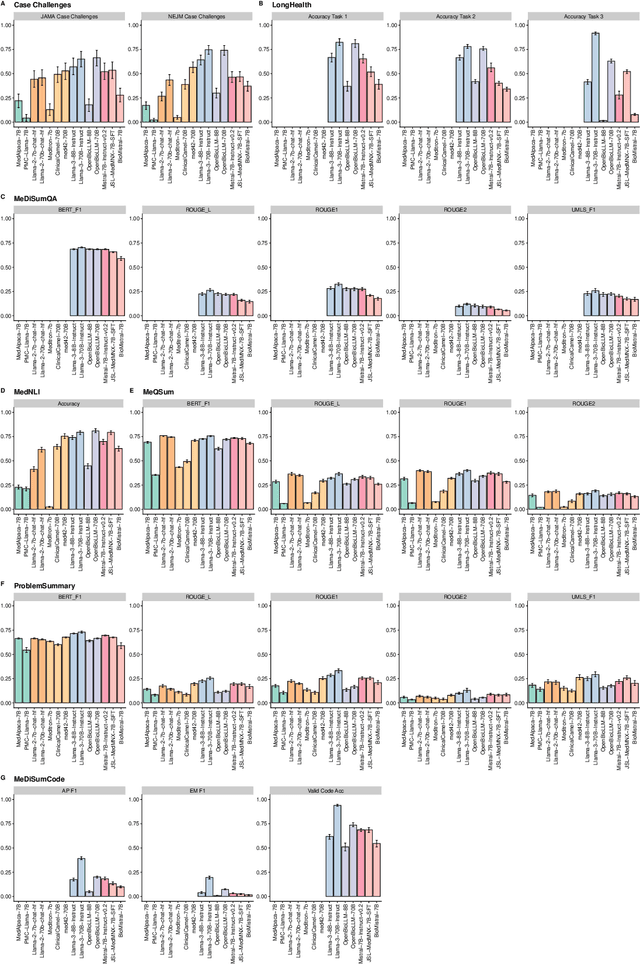
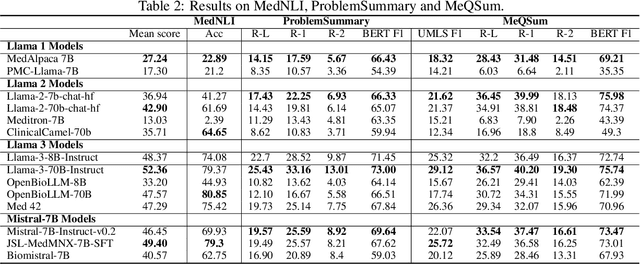
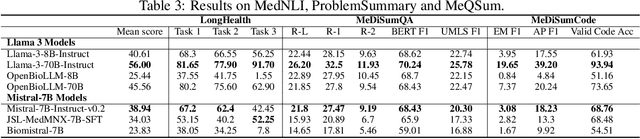
Abstract:Large language models (LLMs) have shown potential in biomedical applications, leading to efforts to fine-tune them on domain-specific data. However, the effectiveness of this approach remains unclear. This study evaluates the performance of biomedically fine-tuned LLMs against their general-purpose counterparts on a variety of clinical tasks. We evaluated their performance on clinical case challenges from the New England Journal of Medicine (NEJM) and the Journal of the American Medical Association (JAMA) and on several clinical tasks (e.g., information extraction, document summarization, and clinical coding). Using benchmarks specifically chosen to be likely outside the fine-tuning datasets of biomedical models, we found that biomedical LLMs mostly perform inferior to their general-purpose counterparts, especially on tasks not focused on medical knowledge. While larger models showed similar performance on case tasks (e.g., OpenBioLLM-70B: 66.4% vs. Llama-3-70B-Instruct: 65% on JAMA cases), smaller biomedical models showed more pronounced underperformance (e.g., OpenBioLLM-8B: 30% vs. Llama-3-8B-Instruct: 64.3% on NEJM cases). Similar trends were observed across the CLUE (Clinical Language Understanding Evaluation) benchmark tasks, with general-purpose models often performing better on text generation, question answering, and coding tasks. Our results suggest that fine-tuning LLMs to biomedical data may not provide the expected benefits and may potentially lead to reduced performance, challenging prevailing assumptions about domain-specific adaptation of LLMs and highlighting the need for more rigorous evaluation frameworks in healthcare AI. Alternative approaches, such as retrieval-augmented generation, may be more effective in enhancing the biomedical capabilities of LLMs without compromising their general knowledge.
Influence of Medical Foreign Bodies on Dark-Field Chest Radiographs: First experiences
Aug 20, 2024Abstract:Objectives: Evaluating the effects and artifacts introduced by medical foreign bodies in clinical dark-field chest radiographs and assessing their influence on the evaluation of pulmonary tissue, compared to conventional radiographs. Material & Methods: This retrospective study analyzed data from subjects enrolled in clinical trials conducted between 2018 and 2021, focusing on chronic obstructive pulmonary disease (COPD) and COVID-19 patients. All patients obtained a radiograph using an in-house developed clinical prototype for grating-based dark-field chest radiography. The prototype simultaneously delivers a conventional and dark-field radiograph. Two radiologists independently assessed the clinical studies to identify patients with foreign bodies. Subsequently, an analysis was conducted on the effects and artifacts attributed to distinct foreign bodies and their impact on the assessment of pulmonary tissue. Results: Overall, 30 subjects with foreign bodies were included in this study (mean age, 64 years +/- 11 [standard deviation]; 15 men). Foreign bodies composed of materials lacking microstructure exhibited a diminished dark-field signal or no discernible signal. Foreign bodies with a microstructure, in our investigations the cementation of the kyphoplasty, produce a positive dark-field signal. Since most foreign bodies lack microstructural features, dark-field imaging revealed fewer signals and artifacts by foreign bodies compared to conventional radiographs. Conclusion: Dark-field radiography enhances the assessment of pulmonary tissue with overlaying foreign bodies compared to conventional radiography. Reduced interfering signals result in fewer overlapping radiopaque artifacts within the investigated regions. This mitigates the impact on image quality and interpretability of the radiographs and the projection-related limitations of radiography compared to CT.
Incorporating Anatomical Awareness for Enhanced Generalizability and Progression Prediction in Deep Learning-Based Radiographic Sacroiliitis Detection
May 12, 2024
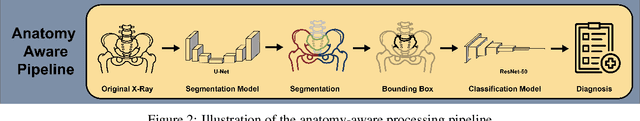

Abstract:Purpose: To examine whether incorporating anatomical awareness into a deep learning model can improve generalizability and enable prediction of disease progression. Methods: This retrospective multicenter study included conventional pelvic radiographs of 4 different patient cohorts focusing on axial spondyloarthritis (axSpA) collected at university and community hospitals. The first cohort, which consisted of 1483 radiographs, was split into training (n=1261) and validation (n=222) sets. The other cohorts comprising 436, 340, and 163 patients, respectively, were used as independent test datasets. For the second cohort, follow-up data of 311 patients was used to examine progression prediction capabilities. Two neural networks were trained, one on images cropped to the bounding box of the sacroiliac joints (anatomy-aware) and the other one on full radiographs. The performance of the models was compared using the area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, and specificity. Results: On the three test datasets, the standard model achieved AUC scores of 0.853, 0.817, 0.947, with an accuracy of 0.770, 0.724, 0.850. Whereas the anatomy-aware model achieved AUC scores of 0.899, 0.846, 0.957, with an accuracy of 0.821, 0.744, 0.906, respectively. The patients who were identified as high risk by the anatomy aware model had an odds ratio of 2.16 (95% CI: 1.19, 3.86) for having progression of radiographic sacroiliitis within 2 years. Conclusion: Anatomical awareness can improve the generalizability of a deep learning model in detecting radiographic sacroiliitis. The model is published as fully open source alongside this study.
MEDBERT.de: A Comprehensive German BERT Model for the Medical Domain
Mar 24, 2023
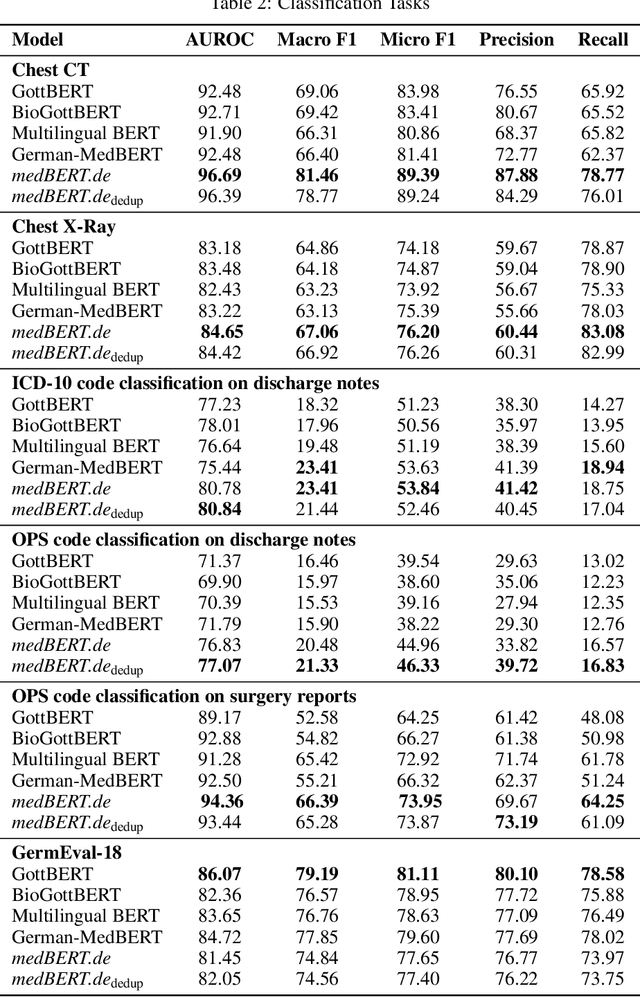
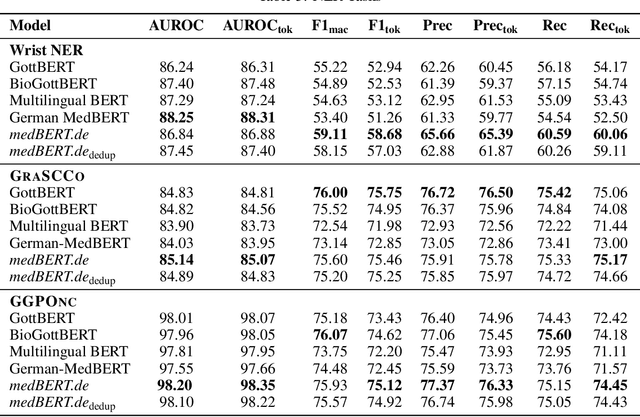
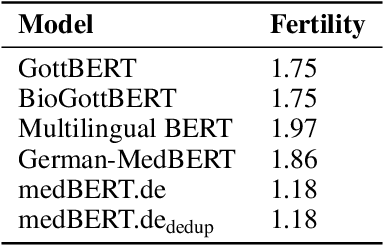
Abstract:This paper presents medBERTde, a pre-trained German BERT model specifically designed for the German medical domain. The model has been trained on a large corpus of 4.7 Million German medical documents and has been shown to achieve new state-of-the-art performance on eight different medical benchmarks covering a wide range of disciplines and medical document types. In addition to evaluating the overall performance of the model, this paper also conducts a more in-depth analysis of its capabilities. We investigate the impact of data deduplication on the model's performance, as well as the potential benefits of using more efficient tokenization methods. Our results indicate that domain-specific models such as medBERTde are particularly useful for longer texts, and that deduplication of training data does not necessarily lead to improved performance. Furthermore, we found that efficient tokenization plays only a minor role in improving model performance, and attribute most of the improved performance to the large amount of training data. To encourage further research, the pre-trained model weights and new benchmarks based on radiological data are made publicly available for use by the scientific community.
What Does DALL-E 2 Know About Radiology?
Sep 27, 2022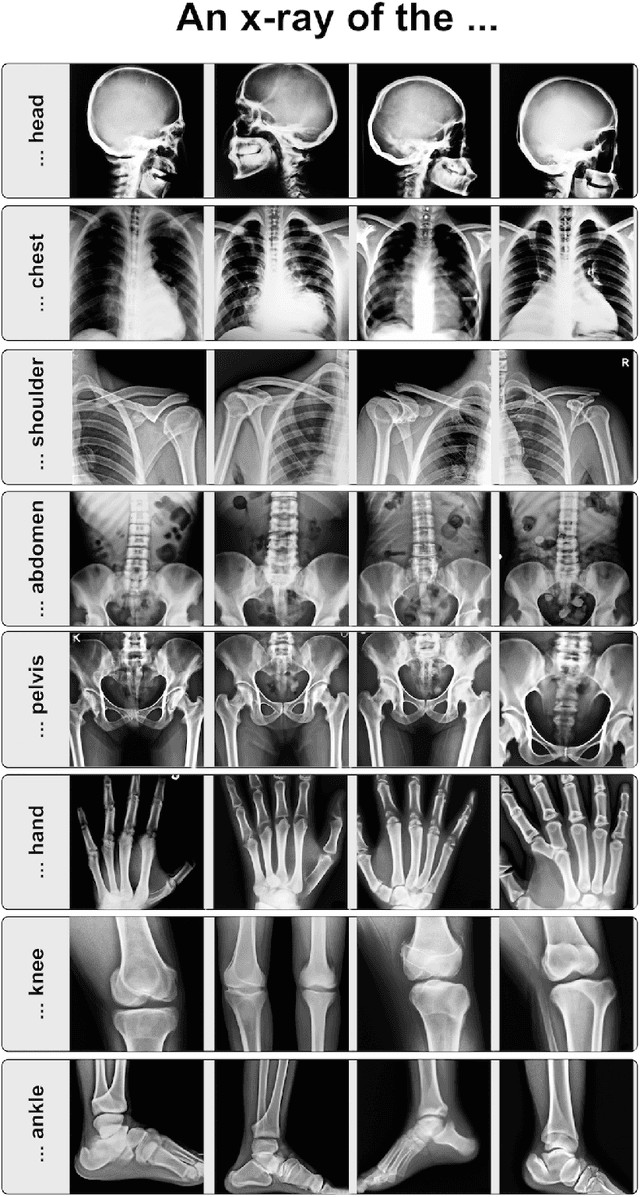
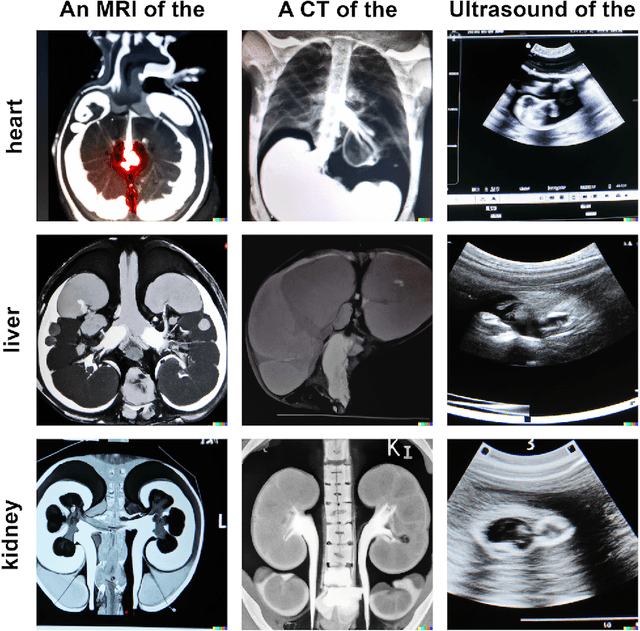
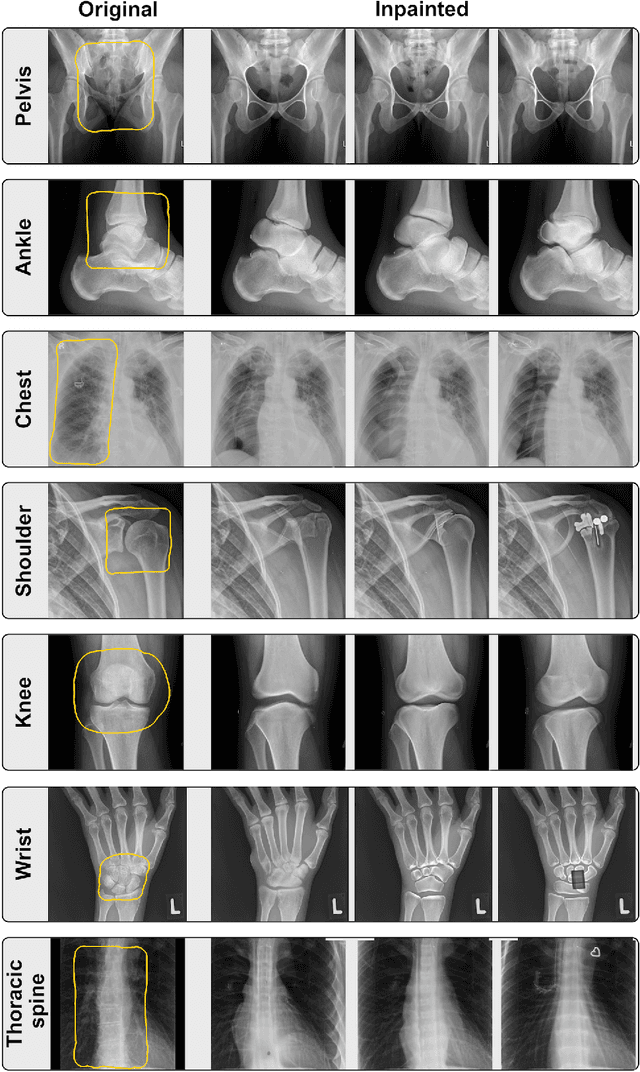
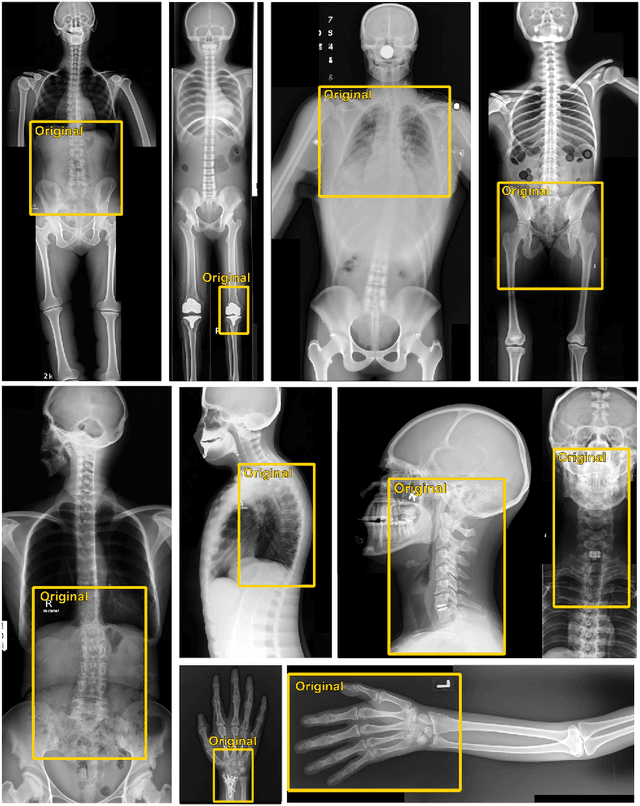
Abstract:Generative models such as DALL-E 2 could represent a promising future tool for image generation, augmentation, and manipulation for artificial intelligence research in radiology provided that these models have sufficient medical domain knowledge. Here we show that DALL-E 2 has learned relevant representations of X-ray images with promising capabilities in terms of zero-shot text-to-image generation of new images, continuation of an image beyond its original boundaries, or removal of elements, while pathology generation or CT, MRI, and ultrasound images are still limited. The use of generative models for augmenting and generating radiological data thus seems feasible, even if further fine-tuning and adaptation of these models to the respective domain is required beforehand.
Per-Pixel Lung Thickness and Lung Capacity Estimation on Chest X-Rays using Convolutional Neural Networks
Oct 27, 2021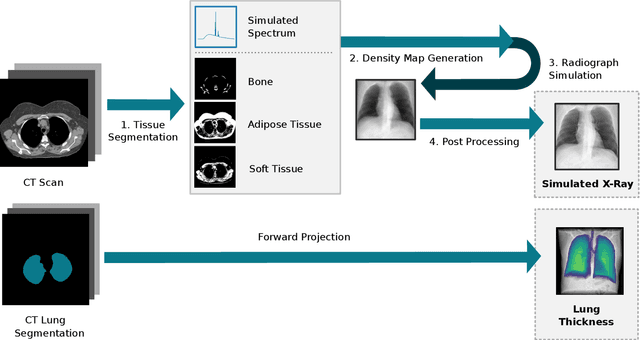
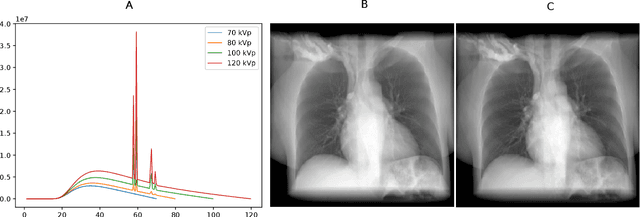
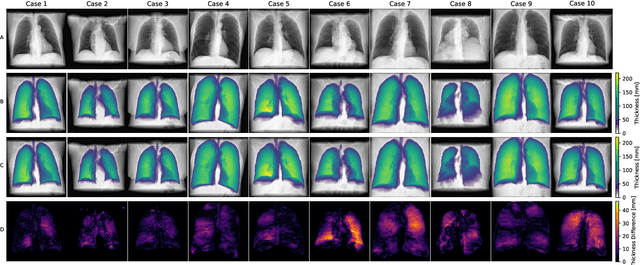
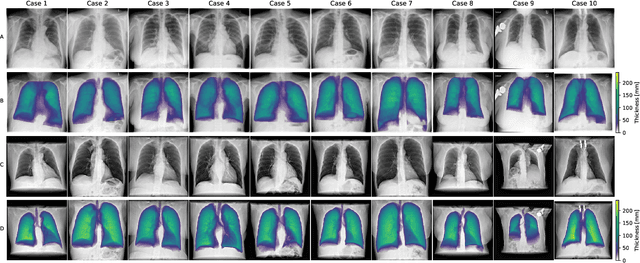
Abstract:Estimating the lung depth on x-ray images could provide both an accurate opportunistic lung volume estimation during clinical routine and improve image contrast in modern structural chest imaging techniques like x-ray dark-field imaging. We present a method based on a convolutional neural network that allows a per-pixel lung thickness estimation and subsequent total lung capacity estimation. The network was trained and validated using 5250 simulated radiographs generated from 525 real CT scans. Furthermore, we are able to infer the model trained with simulation data on real radiographs. For 35 patients, quantitative and qualitative evaluation was performed on standard clinical radiographs. The ground-truth for each patient's total lung volume was defined based on the patients' corresponding CT scan. The mean-absolute error between the estimated lung volume on the 35 real radiographs and groundtruth volume was 0.73 liter. Additionally, we predicted the lung thicknesses on a synthetic dataset of 131 radiographs, where the mean-absolute error was 0.27 liter. The results show, that it is possible to transfer the knowledge obtained in a simulation model to real x-ray images.
Tracked 3D Ultrasound and Deep Neural Network-based Thyroid Segmentation reduce Interobserver Variability in Thyroid Volumetry
Aug 10, 2021

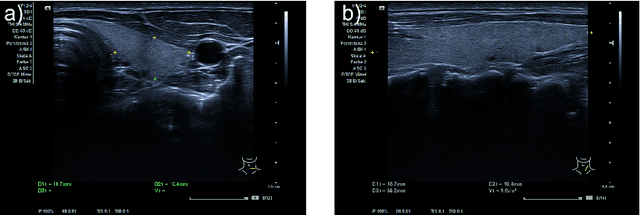
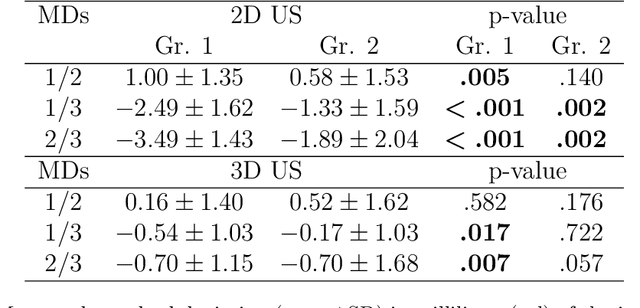
Abstract:Background: Thyroid volumetry is crucial in diagnosis, treatment and monitoring of thyroid diseases. However, conventional thyroid volumetry with 2D ultrasound is highly operator-dependent. This study compares 2D ultrasound and tracked 3D ultrasound with an automatic thyroid segmentation based on a deep neural network regarding inter- and intraobserver variability, time and accuracy. Volume reference was MRI. Methods: 28 healthy volunteers were scanned with 2D and 3D ultrasound as well as by MRI. Three physicians (MD 1, 2, 3) with different levels of experience (6, 4 and 1 a) performed three 2D ultrasound and three tracked 3D ultrasound scans on each volunteer. In the 2D scans the thyroid lobe volumes were calculated with the ellipsoid formula. A convolutional deep neural network (CNN) segmented the 3D thyroid lobes automatically. On MRI (T1 VIBE sequence) the thyroid was manually segmented by an experienced medical doctor. Results: The CNN was trained to obtain a dice score of 0.94. The interobserver variability comparing two MDs showed mean differences for 2D and 3D respectively of 0.58 ml to 0.52 ml (MD1 vs. 2), -1.33 ml to -0.17 ml (MD1 vs. 3) and -1.89 ml to -0.70 ml (MD2 vs. 3). Paired samples t-tests showed significant differences in two comparisons for 2D and none for 3D. Intraobsever variability was similar for 2D and 3D ultrasound. Comparison of ultrasound volumes and MRI volumes by paired samples t-tests showed a significant difference for the 2D volumetry of all MDs, and no significant difference for 3D ultrasound. Acquisition time was significantly shorter for 3D ultrasound. Conclusion: Tracked 3D ultrasound combined with a CNN segmentation significantly reduces interobserver variability in thyroid volumetry and increases the accuracy of the measurements with shorter acquisition times.
NeuralDP Differentially private neural networks by design
Aug 10, 2021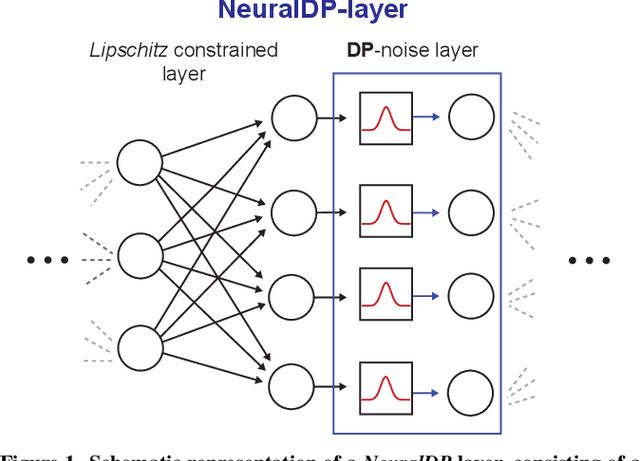
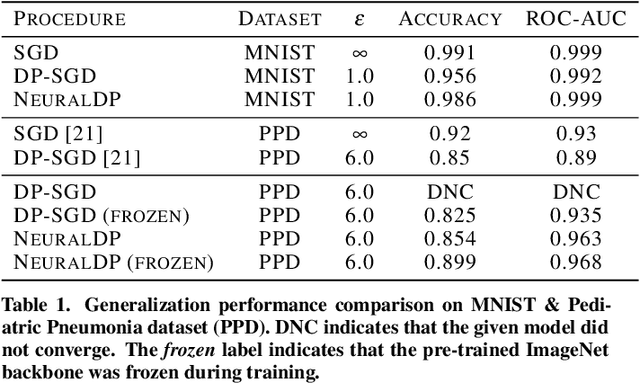
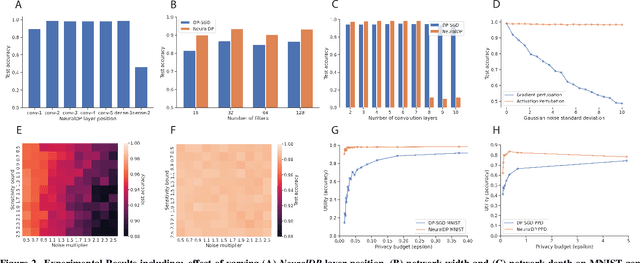
Abstract:The application of differential privacy to the training of deep neural networks holds the promise of allowing large-scale (decentralized) use of sensitive data while providing rigorous privacy guarantees to the individual. The predominant approach to differentially private training of neural networks is DP-SGD, which relies on norm-based gradient clipping as a method for bounding sensitivity, followed by the addition of appropriately calibrated Gaussian noise. In this work we propose NeuralDP, a technique for privatising activations of some layer within a neural network, which by the post-processing properties of differential privacy yields a differentially private network. We experimentally demonstrate on two datasets (MNIST and Pediatric Pneumonia Dataset (PPD)) that our method offers substantially improved privacy-utility trade-offs compared to DP-SGD.
Differentially private training of neural networks with Langevin dynamics for calibrated predictive uncertainty
Aug 04, 2021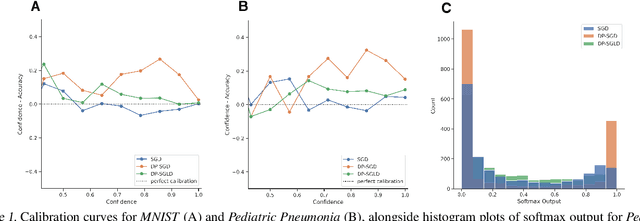
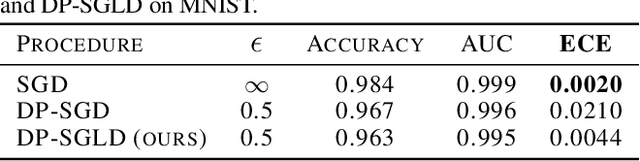
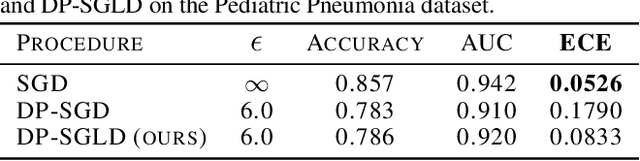
Abstract:We show that differentially private stochastic gradient descent (DP-SGD) can yield poorly calibrated, overconfident deep learning models. This represents a serious issue for safety-critical applications, e.g. in medical diagnosis. We highlight and exploit parallels between stochastic gradient Langevin dynamics, a scalable Bayesian inference technique for training deep neural networks, and DP-SGD, in order to train differentially private, Bayesian neural networks with minor adjustments to the original (DP-SGD) algorithm. Our approach provides considerably more reliable uncertainty estimates than DP-SGD, as demonstrated empirically by a reduction in expected calibration error (MNIST $\sim{5}$-fold, Pediatric Pneumonia Dataset $\sim{2}$-fold).
U-GAT: Multimodal Graph Attention Network for COVID-19 Outcome Prediction
Jul 29, 2021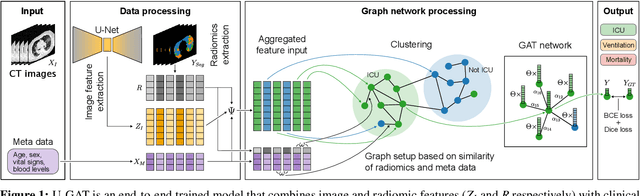
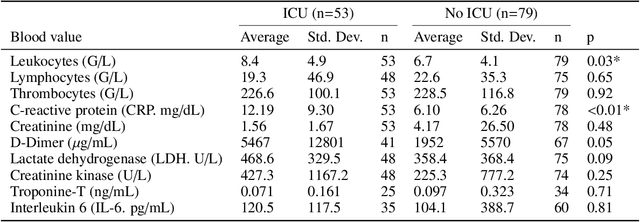
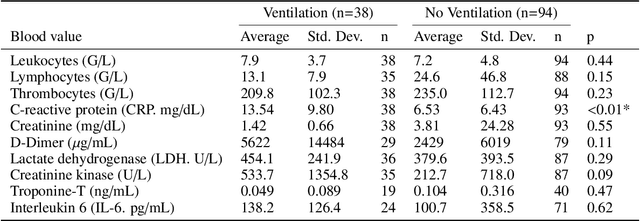
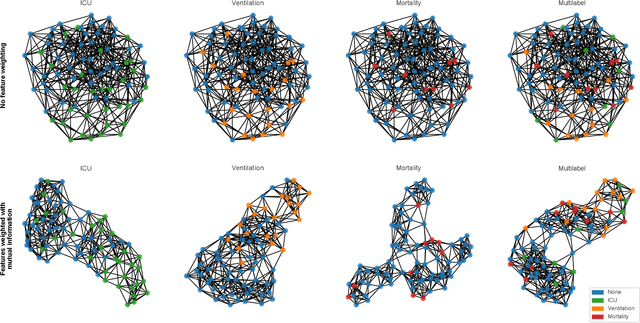
Abstract:During the first wave of COVID-19, hospitals were overwhelmed with the high number of admitted patients. An accurate prediction of the most likely individual disease progression can improve the planning of limited resources and finding the optimal treatment for patients. However, when dealing with a newly emerging disease such as COVID-19, the impact of patient- and disease-specific factors (e.g. body weight or known co-morbidities) on the immediate course of disease is by and large unknown. In the case of COVID-19, the need for intensive care unit (ICU) admission of pneumonia patients is often determined only by acute indicators such as vital signs (e.g. breathing rate, blood oxygen levels), whereas statistical analysis and decision support systems that integrate all of the available data could enable an earlier prognosis. To this end, we propose a holistic graph-based approach combining both imaging and non-imaging information. Specifically, we introduce a multimodal similarity metric to build a population graph for clustering patients and an image-based end-to-end Graph Attention Network to process this graph and predict the COVID-19 patient outcomes: admission to ICU, need for ventilation and mortality. Additionally, the network segments chest CT images as an auxiliary task and extracts image features and radiomics for feature fusion with the available metadata. Results on a dataset collected in Klinikum rechts der Isar in Munich, Germany show that our approach outperforms single modality and non-graph baselines. Moreover, our clustering and graph attention allow for increased understanding of the patient relationships within the population graph and provide insight into the network's decision-making process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge