Markus Krönke
Tracked 3D Ultrasound and Deep Neural Network-based Thyroid Segmentation reduce Interobserver Variability in Thyroid Volumetry
Aug 10, 2021

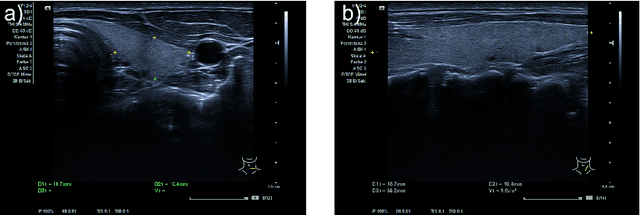
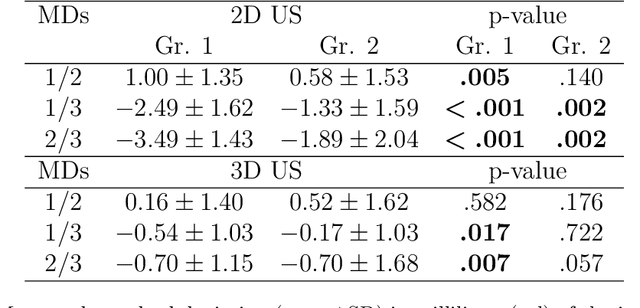
Abstract:Background: Thyroid volumetry is crucial in diagnosis, treatment and monitoring of thyroid diseases. However, conventional thyroid volumetry with 2D ultrasound is highly operator-dependent. This study compares 2D ultrasound and tracked 3D ultrasound with an automatic thyroid segmentation based on a deep neural network regarding inter- and intraobserver variability, time and accuracy. Volume reference was MRI. Methods: 28 healthy volunteers were scanned with 2D and 3D ultrasound as well as by MRI. Three physicians (MD 1, 2, 3) with different levels of experience (6, 4 and 1 a) performed three 2D ultrasound and three tracked 3D ultrasound scans on each volunteer. In the 2D scans the thyroid lobe volumes were calculated with the ellipsoid formula. A convolutional deep neural network (CNN) segmented the 3D thyroid lobes automatically. On MRI (T1 VIBE sequence) the thyroid was manually segmented by an experienced medical doctor. Results: The CNN was trained to obtain a dice score of 0.94. The interobserver variability comparing two MDs showed mean differences for 2D and 3D respectively of 0.58 ml to 0.52 ml (MD1 vs. 2), -1.33 ml to -0.17 ml (MD1 vs. 3) and -1.89 ml to -0.70 ml (MD2 vs. 3). Paired samples t-tests showed significant differences in two comparisons for 2D and none for 3D. Intraobsever variability was similar for 2D and 3D ultrasound. Comparison of ultrasound volumes and MRI volumes by paired samples t-tests showed a significant difference for the 2D volumetry of all MDs, and no significant difference for 3D ultrasound. Acquisition time was significantly shorter for 3D ultrasound. Conclusion: Tracked 3D ultrasound combined with a CNN segmentation significantly reduces interobserver variability in thyroid volumetry and increases the accuracy of the measurements with shorter acquisition times.
End-to-End Learning-Based Ultrasound Reconstruction
Apr 09, 2019


Abstract:Ultrasound imaging is caught between the quest for the highest image quality, and the necessity for clinical usability. Our contribution is two-fold: First, we propose a novel fully convolutional neural network for ultrasound reconstruction. Second, a custom loss function tailored to the modality is employed for end-to-end training of the network. We demonstrate that training a network to map time-delayed raw data to a minimum variance ground truth offers performance increases in a clinical environment. In doing so, a path is explored towards improved clinically viable ultrasound reconstruction. The proposed method displays both promising image reconstruction quality and acquisition frequency when integrated for live ultrasound scanning. A clinical evaluation is conducted to verify the diagnostic usefulness of the proposed method in a clinical setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge