Simultaneous imputation and disease classification in incomplete medical datasets using Multigraph Geometric Matrix Completion (MGMC)
Paper and Code
May 14, 2020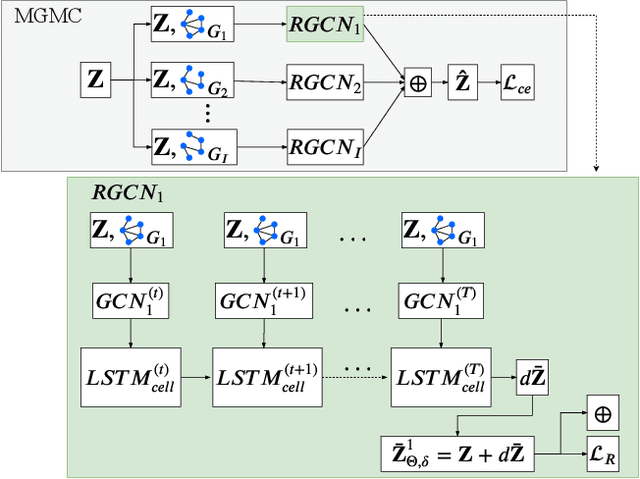
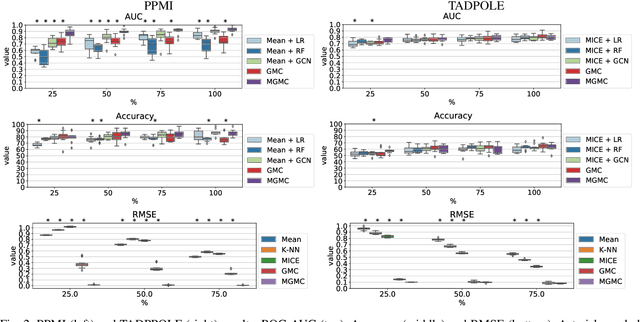
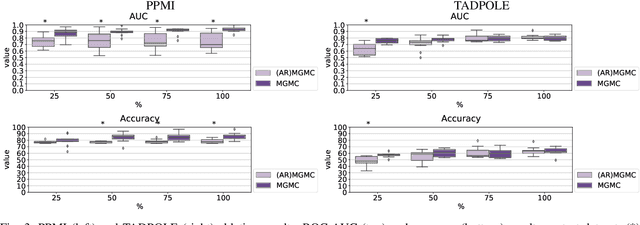
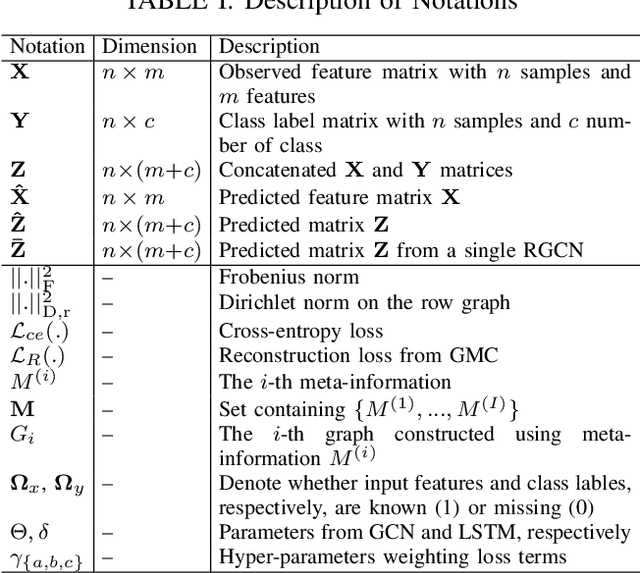
Large-scale population-based studies in medicine are a key resource towards better diagnosis, monitoring, and treatment of diseases. They also serve as enablers of clinical decision support systems, in particular Computer Aided Diagnosis (CADx) using machine learning (ML). Numerous ML approaches for CADx have been proposed in literature. However, these approaches assume full data availability, which is not always feasible in clinical data. To account for missing data, incomplete data samples are either removed or imputed, which could lead to data bias and may negatively affect classification performance. As a solution, we propose an end-to-end learning of imputation and disease prediction of incomplete medical datasets via Multigraph Geometric Matrix Completion (MGMC). MGMC uses multiple recurrent graph convolutional networks, where each graph represents an independent population model based on a key clinical meta-feature like age, sex, or cognitive function. Graph signal aggregation from local patient neighborhoods, combined with multigraph signal fusion via self-attention, has a regularizing effect on both matrix reconstruction and classification performance. Our proposed approach is able to impute class relevant features as well as perform accurate classification on two publicly available medical datasets. We empirically show the superiority of our proposed approach in terms of classification and imputation performance when compared with state-of-the-art approaches. MGMC enables disease prediction in multimodal and incomplete medical datasets. These findings could serve as baseline for future CADx approaches which utilize incomplete datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge