Nikola Milosevic
Stochastic Decision Horizons for Constrained Reinforcement Learning
Feb 04, 2026Abstract:Constrained Markov decision processes (CMDPs) provide a principled model for handling constraints, such as safety and other auxiliary objectives, in reinforcement learning. The common approach of using additive-cost constraints and dual variables often hinders off-policy scalability. We propose a Control as Inference formulation based on stochastic decision horizons, where constraint violations attenuate reward contributions and shorten the effective planning horizon via state-action-dependent continuation. This yields survival-weighted objectives that remain replay-compatible for off-policy actor-critic learning. We propose two violation semantics, absorbing and virtual termination, that share the same survival-weighted return but result in distinct optimization structures that lead to SAC/MPO-style policy improvement. Experiments demonstrate improved sample efficiency and favorable return-violation trade-offs on standard benchmarks. Moreover, MPO with virtual termination (VT-MPO) scales effectively to our high-dimensional musculoskeletal Hyfydy setup.
Physical Embodiment Enables Information Processing Beyond Explicit Sensing in Active Matter
Aug 25, 2025Abstract:Living microorganisms have evolved dedicated sensory machinery to detect environmental perturbations, processing these signals through biochemical networks to guide behavior. Replicating such capabilities in synthetic active matter remains a fundamental challenge. Here, we demonstrate that synthetic active particles can adapt to hidden hydrodynamic perturbations through physical embodiment alone, without explicit sensing mechanisms. Using reinforcement learning to control self-thermophoretic particles, we show that they learn navigation strategies to counteract unobserved flow fields by exploiting information encoded in their physical dynamics. Remarkably, particles successfully navigate perturbations that are not included in their state inputs, revealing that embodied dynamics can serve as an implicit sensing mechanism. This discovery establishes physical embodiment as a computational resource for information processing in active matter, with implications for autonomous microrobotic systems and bio-inspired computation.
Embedding Safety into RL: A New Take on Trust Region Methods
Nov 05, 2024



Abstract:Reinforcement Learning (RL) agents are able to solve a wide variety of tasks but are prone to producing unsafe behaviors. Constrained Markov Decision Processes (CMDPs) provide a popular framework for incorporating safety constraints. However, common solution methods often compromise reward maximization by being overly conservative or allow unsafe behavior during training. We propose Constrained Trust Region Policy Optimization (C-TRPO), a novel approach that modifies the geometry of the policy space based on the safety constraints and yields trust regions composed exclusively of safe policies, ensuring constraint satisfaction throughout training. We theoretically study the convergence and update properties of C-TRPO and highlight connections to TRPO, Natural Policy Gradient (NPG), and Constrained Policy Optimization (CPO). Finally, we demonstrate experimentally that C-TRPO significantly reduces constraint violations while achieving competitive reward maximization compared to state-of-the-art CMDP algorithms.
Fact Finder -- Enhancing Domain Expertise of Large Language Models by Incorporating Knowledge Graphs
Aug 06, 2024



Abstract:Recent advancements in Large Language Models (LLMs) have showcased their proficiency in answering natural language queries. However, their effectiveness is hindered by limited domain-specific knowledge, raising concerns about the reliability of their responses. We introduce a hybrid system that augments LLMs with domain-specific knowledge graphs (KGs), thereby aiming to enhance factual correctness using a KG-based retrieval approach. We focus on a medical KG to demonstrate our methodology, which includes (1) pre-processing, (2) Cypher query generation, (3) Cypher query processing, (4) KG retrieval, and (5) LLM-enhanced response generation. We evaluate our system on a curated dataset of 69 samples, achieving a precision of 78\% in retrieving correct KG nodes. Our findings indicate that the hybrid system surpasses a standalone LLM in accuracy and completeness, as verified by an LLM-as-a-Judge evaluation method. This positions the system as a promising tool for applications that demand factual correctness and completeness, such as target identification -- a critical process in pinpointing biological entities for disease treatment or crop enhancement. Moreover, its intuitive search interface and ability to provide accurate responses within seconds make it well-suited for time-sensitive, precision-focused research contexts. We publish the source code together with the dataset and the prompt templates used.
Open Problem: Active Representation Learning
Jun 06, 2024
Abstract:In this work, we introduce the concept of Active Representation Learning, a novel class of problems that intertwines exploration and representation learning within partially observable environments. We extend ideas from Active Simultaneous Localization and Mapping (active SLAM), and translate them to scientific discovery problems, exemplified by adaptive microscopy. We explore the need for a framework that derives exploration skills from representations that are in some sense actionable, aiming to enhance the efficiency and effectiveness of data collection and model building in the natural sciences.
Relationship extraction for knowledge graph creation from biomedical literature
Jan 05, 2022



Abstract:Biomedical research is growing in such an exponential pace that scientists, researchers and practitioners are no more able to cope with the amount of published literature in the domain. The knowledge presented in the literature needs to be systematized in such a ways that claims and hypothesis can be easily found, accessed and validated. Knowledge graphs can provide such framework for semantic knowledge representation from literature. However, in order to build knowledge graph, it is necessary to extract knowledge in form of relationships between biomedical entities and normalize both entities and relationship types. In this paper, we present and compare few rule-based and machine learning-based (Naive Bayes, Random Forests as examples of traditional machine learning methods and T5-based model as an example of modern deep learning) methods for scalable relationship extraction from biomedical literature for the integration into the knowledge graphs. We examine how resilient are these various methods to unbalanced and fairly small datasets, showing that T5 model handles well both small datasets, due to its pre-training on large C4 dataset as well as unbalanced data. The best performing model was T5 model fine-tuned on balanced data, with reported F1-score of 0.88.
MASK: A flexible framework to facilitate de-identification of clinical texts
May 24, 2020



Abstract:Medical health records and clinical summaries contain a vast amount of important information in textual form that can help advancing research on treatments, drugs and public health. However, the majority of these information is not shared because they contain private information about patients, their families, or medical staff treating them. Regulations such as HIPPA in the US, PHIPPA in Canada and GDPR regulate the protection, processing and distribution of this information. In case this information is de-identified and personal information are replaced or redacted, they could be distributed to the research community. In this paper, we present MASK, a software package that is designed to perform the de-identification task. The software is able to perform named entity recognition using some of the state-of-the-art techniques and then mask or redact recognized entities. The user is able to select named entity recognition algorithm (currently implemented are two versions of CRF-based techniques and BiLSTM-based neural network with pre-trained GLoVe and ELMo embedding) and masking algorithm (e.g. shift dates, replace names/locations, totally redact entity).
Deep learning guided Android malware and anomaly detection
Oct 23, 2019



Abstract:In the past decade, the cyber-crime related to mobile devices has increased. Mobile devices, especially the ones running on Android operating system are particularly interesting to malware creators, as the users often keep the biggest amount of personal information on their mobile devices, such as their contacts, social media profiles, emails, and bank accounts. Both dynamic and static malware analysis is necessary to prevent and detect malware, as both techniques have their benefits and shortcomings. In this paper, we propose a deep learning technique that relies on LSTM and encoder-decoder neural network architectures for dynamic malware analysis based on CPU, memory and battery usage. The proposed system is able to detect and notify users about anomalies in system that is likely consequence of malware behaviour. The method was implemented as a part of OWASP Seraphimdroids anti-malware mechanism and notifies users about anomalies on their devices. The method proved to perform with an F1-score of 79.2%.
GNTeam at 2018 n2c2: Feature-augmented BiLSTM-CRF for drug-related entity recognition in hospital discharge summaries
Sep 23, 2019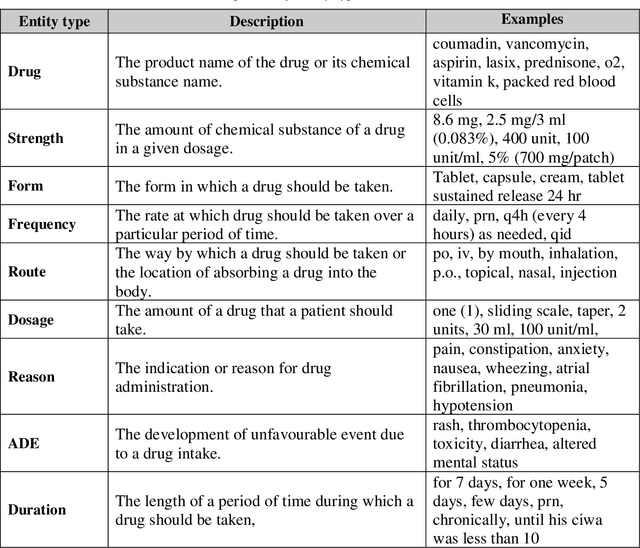
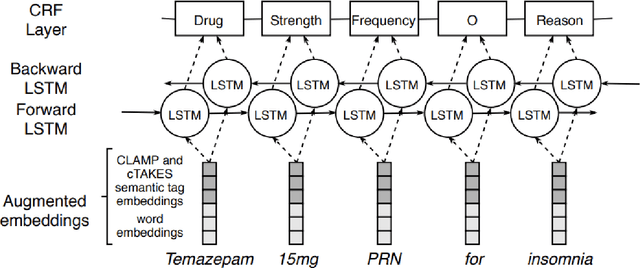

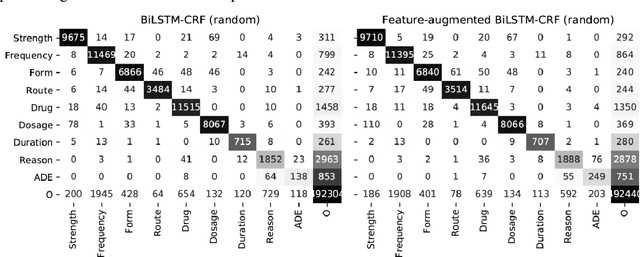
Abstract:Monitoring the administration of drugs and adverse drug reactions are key parts of pharmacovigilance. In this paper, we explore the extraction of drug mentions and drug-related information (reason for taking a drug, route, frequency, dosage, strength, form, duration, and adverse events) from hospital discharge summaries through deep learning that relies on various representations for clinical named entity recognition. This work was officially part of the 2018 n2c2 shared task, and we use the data supplied as part of the task. We developed two deep learning architecture based on recurrent neural networks and pre-trained language models. We also explore the effect of augmenting word representations with semantic features for clinical named entity recognition. Our feature-augmented BiLSTM-CRF model performed with F1-score of 92.67% and ranked 4th for entity extraction sub-task among submitted systems to n2c2 challenge. The recurrent neural networks that use the pre-trained domain-specific word embeddings and a CRF layer for label optimization perform drug, adverse event and related entities extraction with micro-averaged F1-score of over 91%. The augmentation of word vectors with semantic features extracted using available clinical NLP toolkits can further improve the performance. Word embeddings that are pre-trained on a large unannotated corpus of relevant documents and further fine-tuned to the task perform rather well. However, the augmentation of word embeddings with semantic features can help improve the performance (primarily by boosting precision) of drug-related named entity recognition from electronic health records.
Extracting adverse drug reactions and their context using sequence labelling ensembles in TAC2017
May 28, 2019


Abstract:Adverse drug reactions (ADRs) are unwanted or harmful effects experienced after the administration of a certain drug or a combination of drugs, presenting a challenge for drug development and drug administration. In this paper, we present a set of taggers for extracting adverse drug reactions and related entities, including factors, severity, negations, drug class and animal. The systems used a mix of rule-based, machine learning (CRF) and deep learning (BLSTM with word2vec embeddings) methodologies in order to annotate the data. The systems were submitted to adverse drug reaction shared task, organised during Text Analytics Conference in 2017 by National Institute for Standards and Technology, archiving F1-scores of 76.00 and 75.61 respectively.
* Paper describing submission for TAC ADR shared task
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge