Maksim Belousov
GNTeam at 2018 n2c2: Feature-augmented BiLSTM-CRF for drug-related entity recognition in hospital discharge summaries
Sep 23, 2019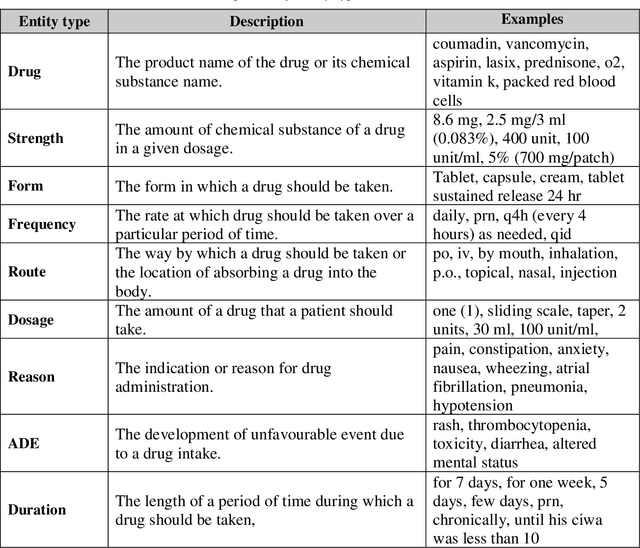
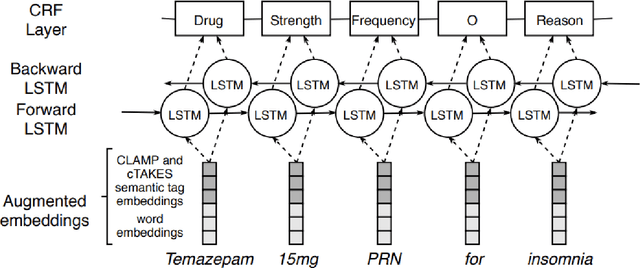

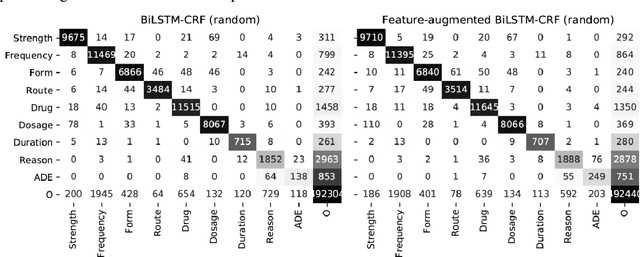
Abstract:Monitoring the administration of drugs and adverse drug reactions are key parts of pharmacovigilance. In this paper, we explore the extraction of drug mentions and drug-related information (reason for taking a drug, route, frequency, dosage, strength, form, duration, and adverse events) from hospital discharge summaries through deep learning that relies on various representations for clinical named entity recognition. This work was officially part of the 2018 n2c2 shared task, and we use the data supplied as part of the task. We developed two deep learning architecture based on recurrent neural networks and pre-trained language models. We also explore the effect of augmenting word representations with semantic features for clinical named entity recognition. Our feature-augmented BiLSTM-CRF model performed with F1-score of 92.67% and ranked 4th for entity extraction sub-task among submitted systems to n2c2 challenge. The recurrent neural networks that use the pre-trained domain-specific word embeddings and a CRF layer for label optimization perform drug, adverse event and related entities extraction with micro-averaged F1-score of over 91%. The augmentation of word vectors with semantic features extracted using available clinical NLP toolkits can further improve the performance. Word embeddings that are pre-trained on a large unannotated corpus of relevant documents and further fine-tuned to the task perform rather well. However, the augmentation of word embeddings with semantic features can help improve the performance (primarily by boosting precision) of drug-related named entity recognition from electronic health records.
Extracting adverse drug reactions and their context using sequence labelling ensembles in TAC2017
May 28, 2019


Abstract:Adverse drug reactions (ADRs) are unwanted or harmful effects experienced after the administration of a certain drug or a combination of drugs, presenting a challenge for drug development and drug administration. In this paper, we present a set of taggers for extracting adverse drug reactions and related entities, including factors, severity, negations, drug class and animal. The systems used a mix of rule-based, machine learning (CRF) and deep learning (BLSTM with word2vec embeddings) methodologies in order to annotate the data. The systems were submitted to adverse drug reaction shared task, organised during Text Analytics Conference in 2017 by National Institute for Standards and Technology, archiving F1-scores of 76.00 and 75.61 respectively.
* Paper describing submission for TAC ADR shared task
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge